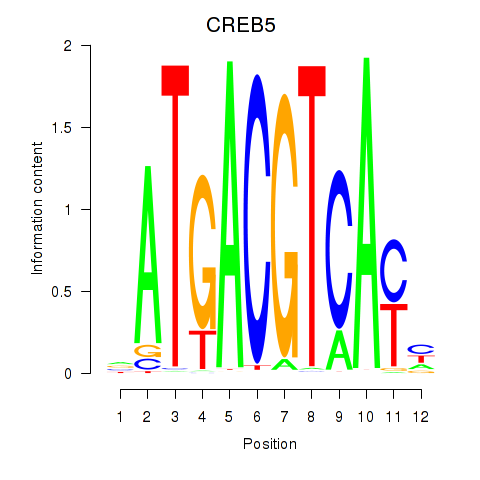
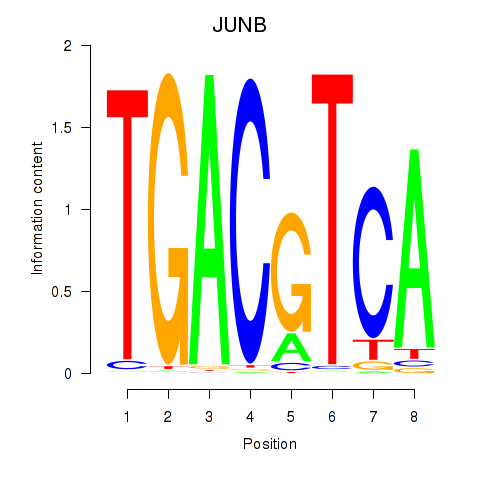
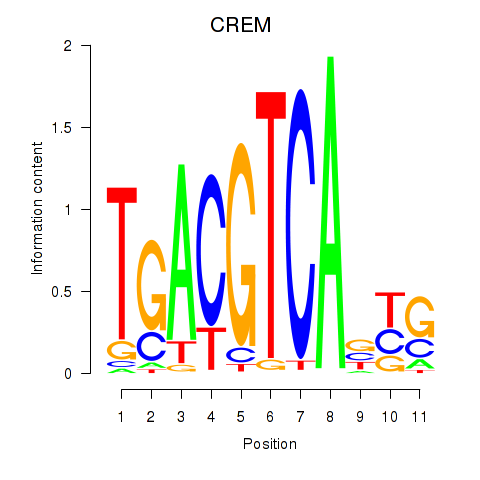
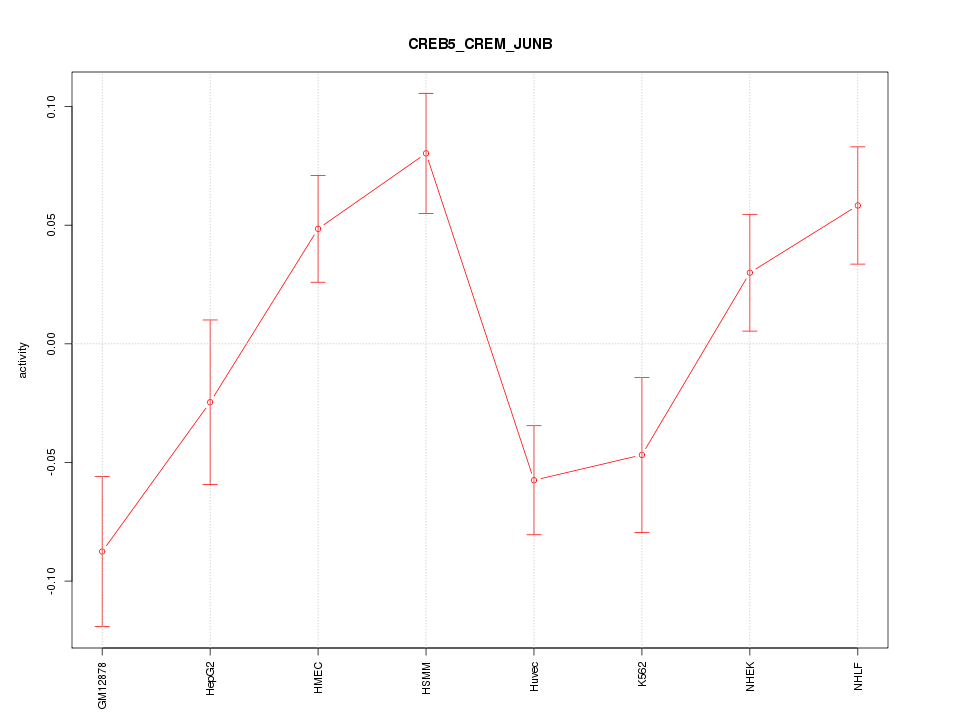
| 1.3 |
4.0 |
GO:0070886 |
positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.9 |
18.4 |
GO:0035329 |
hippo signaling(GO:0035329) |
| 0.9 |
5.1 |
GO:0006931 |
substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.8 |
2.3 |
GO:0043276 |
anoikis(GO:0043276) |
| 0.5 |
8.0 |
GO:0034446 |
substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.5 |
1.4 |
GO:0010966 |
regulation of phosphate transport(GO:0010966) |
| 0.4 |
1.3 |
GO:0032988 |
ribonucleoprotein complex disassembly(GO:0032988) |
| 0.4 |
4.5 |
GO:0033327 |
Leydig cell differentiation(GO:0033327) |
| 0.4 |
1.1 |
GO:0014028 |
notochord formation(GO:0014028) axial mesoderm morphogenesis(GO:0048319) |
| 0.4 |
1.1 |
GO:0051365 |
cellular response to potassium ion starvation(GO:0051365) |
| 0.4 |
1.1 |
GO:0072677 |
eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.3 |
9.0 |
GO:0042730 |
fibrinolysis(GO:0042730) |
| 0.3 |
2.0 |
GO:0021520 |
spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.3 |
5.3 |
GO:0050812 |
acetyl-CoA biosynthetic process from pyruvate(GO:0006086) regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.3 |
1.5 |
GO:0008588 |
release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.3 |
3.2 |
GO:0001706 |
endoderm formation(GO:0001706) |
| 0.3 |
0.8 |
GO:0071277 |
cellular response to calcium ion(GO:0071277) |
| 0.3 |
3.3 |
GO:0055015 |
ventricular cardiac muscle cell development(GO:0055015) |
| 0.3 |
2.3 |
GO:0010799 |
regulation of peptidyl-threonine phosphorylation(GO:0010799) positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.2 |
0.7 |
GO:0090086 |
negative regulation of protein deubiquitination(GO:0090086) |
| 0.2 |
1.9 |
GO:0046325 |
negative regulation of glucose import(GO:0046325) |
| 0.2 |
7.0 |
GO:0031572 |
G2 DNA damage checkpoint(GO:0031572) |
| 0.2 |
0.7 |
GO:0060370 |
susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.2 |
1.2 |
GO:0071392 |
cellular response to estradiol stimulus(GO:0071392) |
| 0.2 |
1.2 |
GO:0031573 |
intra-S DNA damage checkpoint(GO:0031573) |
| 0.2 |
4.0 |
GO:0032012 |
regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 |
1.8 |
GO:0002553 |
histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.2 |
0.6 |
GO:0002318 |
myeloid progenitor cell differentiation(GO:0002318) |
| 0.2 |
2.0 |
GO:0031268 |
pseudopodium organization(GO:0031268) |
| 0.2 |
1.3 |
GO:0042989 |
sequestering of actin monomers(GO:0042989) |
| 0.2 |
0.5 |
GO:0014022 |
neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.2 |
7.9 |
GO:0045740 |
positive regulation of DNA replication(GO:0045740) |
| 0.2 |
5.7 |
GO:0006730 |
one-carbon metabolic process(GO:0006730) |
| 0.2 |
0.8 |
GO:0001661 |
conditioned taste aversion(GO:0001661) |
| 0.2 |
3.0 |
GO:0014003 |
oligodendrocyte development(GO:0014003) |
| 0.1 |
0.6 |
GO:0010332 |
response to gamma radiation(GO:0010332) |
| 0.1 |
0.7 |
GO:0007182 |
common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.1 |
0.4 |
GO:0001544 |
initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.1 |
1.1 |
GO:0051294 |
establishment of mitotic spindle orientation(GO:0000132) establishment of spindle orientation(GO:0051294) |
| 0.1 |
1.9 |
GO:0043508 |
negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 |
1.5 |
GO:0046839 |
phospholipid dephosphorylation(GO:0046839) |
| 0.1 |
1.6 |
GO:0009651 |
response to salt stress(GO:0009651) |
| 0.1 |
0.5 |
GO:0072385 |
minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.1 |
0.3 |
GO:0007000 |
nucleolus organization(GO:0007000) |
| 0.1 |
0.4 |
GO:0070527 |
platelet aggregation(GO:0070527) |
| 0.1 |
0.3 |
GO:0009396 |
folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.1 |
0.4 |
GO:0051877 |
pigment granule aggregation in cell center(GO:0051877) |
| 0.1 |
0.7 |
GO:0010713 |
negative regulation of collagen metabolic process(GO:0010713) negative regulation of collagen biosynthetic process(GO:0032966) negative regulation of multicellular organismal metabolic process(GO:0044252) |
| 0.1 |
0.4 |
GO:0071480 |
cellular response to gamma radiation(GO:0071480) |
| 0.1 |
0.2 |
GO:0035120 |
post-embryonic appendage morphogenesis(GO:0035120) |
| 0.1 |
1.3 |
GO:0030212 |
hyaluronan metabolic process(GO:0030212) |
| 0.1 |
1.3 |
GO:0006657 |
CDP-choline pathway(GO:0006657) |
| 0.1 |
1.3 |
GO:0030854 |
positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 |
0.3 |
GO:0046604 |
regulation of mitotic centrosome separation(GO:0046602) positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.1 |
0.2 |
GO:0060689 |
cell differentiation involved in salivary gland development(GO:0060689) |
| 0.1 |
0.7 |
GO:0019321 |
pentose metabolic process(GO:0019321) |
| 0.1 |
3.3 |
GO:0007368 |
determination of left/right symmetry(GO:0007368) |
| 0.1 |
0.3 |
GO:0000389 |
mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 |
5.9 |
GO:0030049 |
muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.1 |
0.1 |
GO:0010837 |
regulation of keratinocyte proliferation(GO:0010837) |
| 0.1 |
0.1 |
GO:0033484 |
nitric oxide homeostasis(GO:0033484) |
| 0.1 |
1.8 |
GO:0034199 |
activation of protein kinase A activity(GO:0034199) |
| 0.1 |
2.0 |
GO:0015985 |
energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 |
0.3 |
GO:0010508 |
positive regulation of autophagy(GO:0010508) |
| 0.1 |
1.9 |
GO:0044349 |
nucleotide-excision repair, DNA damage removal(GO:0000718) DNA excision(GO:0044349) |
| 0.1 |
1.1 |
GO:0051281 |
positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) |
| 0.1 |
0.6 |
GO:0051014 |
actin filament severing(GO:0051014) |
| 0.1 |
0.2 |
GO:0043163 |
cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.1 |
0.9 |
GO:0007567 |
parturition(GO:0007567) |
| 0.1 |
2.9 |
GO:0070936 |
protein K48-linked ubiquitination(GO:0070936) |
| 0.1 |
1.1 |
GO:0035196 |
production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.1 |
0.2 |
GO:0032650 |
regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) |
| 0.1 |
0.1 |
GO:0045760 |
positive regulation of action potential(GO:0045760) |
| 0.0 |
0.7 |
GO:0008634 |
obsolete negative regulation of survival gene product expression(GO:0008634) |
| 0.0 |
0.4 |
GO:0071692 |
sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 |
1.8 |
GO:0007026 |
negative regulation of microtubule depolymerization(GO:0007026) regulation of microtubule depolymerization(GO:0031114) |
| 0.0 |
0.4 |
GO:0010815 |
bradykinin catabolic process(GO:0010815) |
| 0.0 |
0.4 |
GO:0001967 |
suckling behavior(GO:0001967) |
| 0.0 |
2.3 |
GO:0007043 |
cell-cell junction assembly(GO:0007043) |
| 0.0 |
0.6 |
GO:0060644 |
mammary gland epithelial cell differentiation(GO:0060644) |
| 0.0 |
0.1 |
GO:0010792 |
DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 |
0.4 |
GO:0015014 |
heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 |
1.1 |
GO:0031424 |
keratinization(GO:0031424) |
| 0.0 |
3.3 |
GO:0009408 |
response to heat(GO:0009408) |
| 0.0 |
7.4 |
GO:0007015 |
actin filament organization(GO:0007015) |
| 0.0 |
0.1 |
GO:0014831 |
intestine smooth muscle contraction(GO:0014827) gastro-intestinal system smooth muscle contraction(GO:0014831) |
| 0.0 |
0.3 |
GO:0006622 |
protein targeting to lysosome(GO:0006622) protein targeting to vacuole(GO:0006623) protein localization to lysosome(GO:0061462) protein localization to vacuole(GO:0072665) establishment of protein localization to vacuole(GO:0072666) |
| 0.0 |
0.3 |
GO:0042797 |
5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 |
0.1 |
GO:0008628 |
hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.0 |
2.7 |
GO:0007588 |
excretion(GO:0007588) |
| 0.0 |
0.2 |
GO:0007341 |
penetration of zona pellucida(GO:0007341) |
| 0.0 |
0.3 |
GO:0007185 |
transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 |
0.6 |
GO:0045104 |
intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 |
0.1 |
GO:0042416 |
dopamine biosynthetic process(GO:0042416) |
| 0.0 |
0.2 |
GO:0042149 |
cellular response to glucose starvation(GO:0042149) |
| 0.0 |
0.2 |
GO:0006610 |
ribosomal protein import into nucleus(GO:0006610) |
| 0.0 |
0.3 |
GO:0033523 |
histone monoubiquitination(GO:0010390) histone H2B ubiquitination(GO:0033523) |
| 0.0 |
0.1 |
GO:0070935 |
3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 |
2.2 |
GO:0007623 |
circadian rhythm(GO:0007623) |
| 0.0 |
0.2 |
GO:0021955 |
central nervous system projection neuron axonogenesis(GO:0021952) central nervous system neuron axonogenesis(GO:0021955) |
| 0.0 |
7.4 |
GO:0007517 |
muscle organ development(GO:0007517) |
| 0.0 |
0.4 |
GO:0032456 |
endocytic recycling(GO:0032456) |
| 0.0 |
0.1 |
GO:0042518 |
negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) negative regulation of tyrosine phosphorylation of STAT protein(GO:0042532) |
| 0.0 |
0.3 |
GO:0006376 |
mRNA splice site selection(GO:0006376) |
| 0.0 |
0.3 |
GO:0048268 |
negative regulation of receptor-mediated endocytosis(GO:0048261) clathrin coat assembly(GO:0048268) |
| 0.0 |
0.9 |
GO:0030520 |
intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.0 |
0.4 |
GO:0007172 |
signal complex assembly(GO:0007172) |
| 0.0 |
1.9 |
GO:0006368 |
transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 |
0.1 |
GO:0021535 |
cell migration in hindbrain(GO:0021535) |
| 0.0 |
0.2 |
GO:0050847 |
progesterone receptor signaling pathway(GO:0050847) |
| 0.0 |
0.6 |
GO:0043473 |
pigmentation(GO:0043473) |
| 0.0 |
0.1 |
GO:0051005 |
negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 |
0.1 |
GO:0007616 |
long-term memory(GO:0007616) synaptic vesicle docking(GO:0016081) calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 |
0.0 |
GO:0010761 |
fibroblast migration(GO:0010761) |
| 0.0 |
2.3 |
GO:0000187 |
activation of MAPK activity(GO:0000187) |
| 0.0 |
0.2 |
GO:0060765 |
regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 |
0.3 |
GO:0043507 |
positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 |
1.0 |
GO:0006914 |
autophagy(GO:0006914) |
| 0.0 |
0.2 |
GO:0090083 |
regulation of inclusion body assembly(GO:0090083) |
| 0.0 |
0.0 |
GO:0046122 |
purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.0 |
1.4 |
GO:0019722 |
calcium-mediated signaling(GO:0019722) |
| 0.0 |
0.5 |
GO:0006699 |
bile acid biosynthetic process(GO:0006699) |
| 0.0 |
0.6 |
GO:0015914 |
phospholipid transport(GO:0015914) |
| 0.0 |
0.1 |
GO:0043982 |
histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 |
0.1 |
GO:0030813 |
positive regulation of nucleotide catabolic process(GO:0030813) positive regulation of glycolytic process(GO:0045821) positive regulation of nucleoside metabolic process(GO:0045979) positive regulation of cofactor metabolic process(GO:0051194) positive regulation of coenzyme metabolic process(GO:0051197) positive regulation of ATP metabolic process(GO:1903580) |
| 0.0 |
0.1 |
GO:0051445 |
regulation of meiotic nuclear division(GO:0040020) regulation of meiotic cell cycle(GO:0051445) |
| 0.0 |
0.4 |
GO:0046464 |
triglyceride catabolic process(GO:0019433) neutral lipid catabolic process(GO:0046461) acylglycerol catabolic process(GO:0046464) |
| 0.0 |
0.1 |
GO:0071372 |
response to light intensity(GO:0009642) response to follicle-stimulating hormone(GO:0032354) cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 |
0.3 |
GO:0007413 |
axonal fasciculation(GO:0007413) |
| 0.0 |
0.1 |
GO:0006398 |
mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 |
0.3 |
GO:0051928 |
positive regulation of calcium ion transport(GO:0051928) |
| 0.0 |
0.5 |
GO:0050931 |
melanocyte differentiation(GO:0030318) pigment cell differentiation(GO:0050931) |
| 0.0 |
0.1 |
GO:0016574 |
histone ubiquitination(GO:0016574) |
| 0.0 |
0.1 |
GO:0070536 |
protein K63-linked deubiquitination(GO:0070536) |
| 0.0 |
0.2 |
GO:0050690 |
regulation of defense response to virus by virus(GO:0050690) |