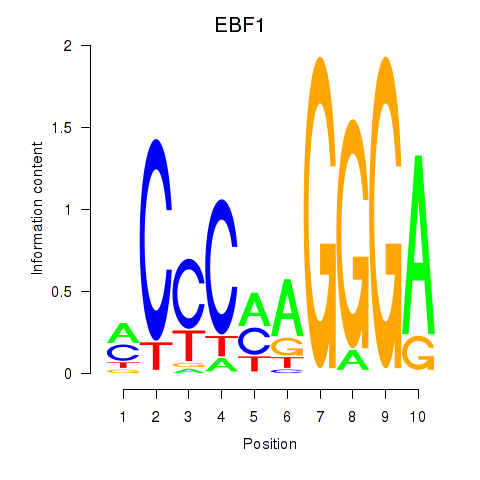
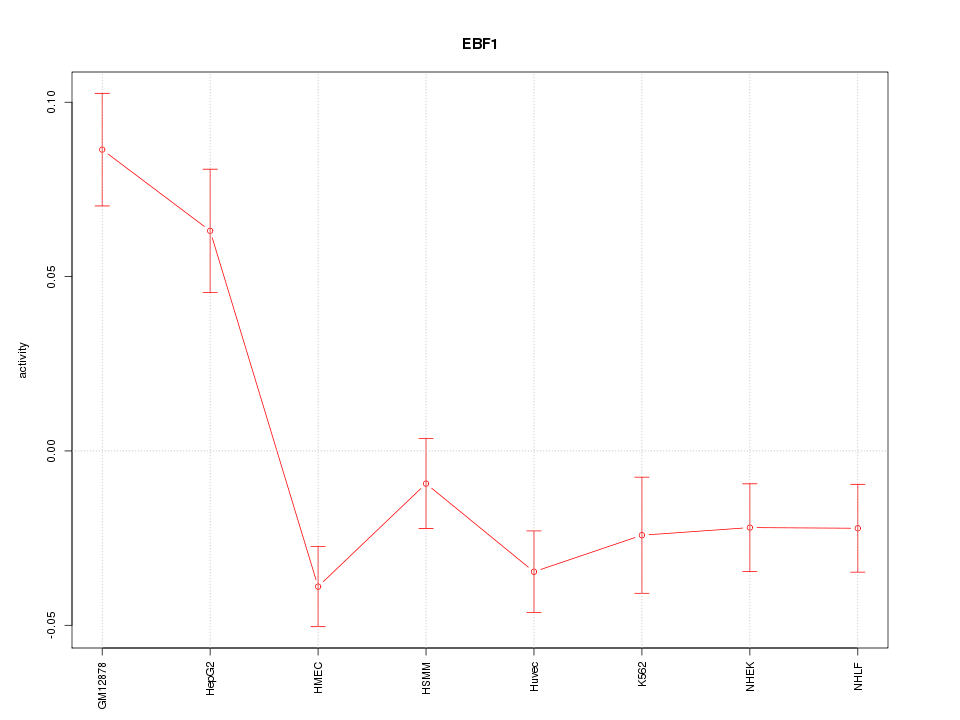
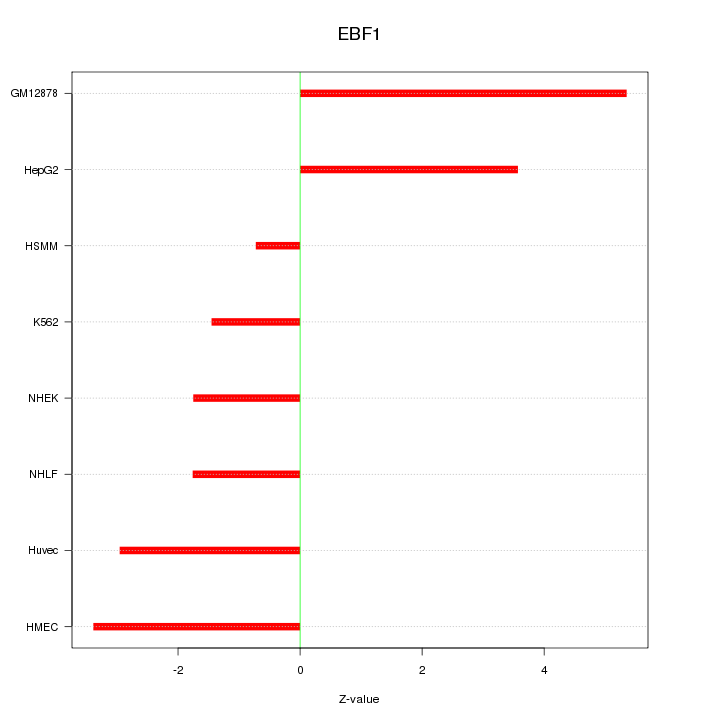
| 2.4 |
9.5 |
GO:0010903 |
negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) |
| 1.4 |
4.1 |
GO:0019886 |
antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 1.2 |
3.5 |
GO:1903961 |
positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) positive regulation of anion transmembrane transport(GO:1903961) |
| 1.1 |
3.4 |
GO:0046210 |
nitric oxide catabolic process(GO:0046210) |
| 0.9 |
6.9 |
GO:0000185 |
activation of MAPKKK activity(GO:0000185) |
| 0.8 |
3.0 |
GO:1904729 |
regulation of intestinal cholesterol absorption(GO:0030300) regulation of intestinal absorption(GO:1904478) regulation of intestinal lipid absorption(GO:1904729) |
| 0.6 |
2.6 |
GO:0051712 |
positive regulation of killing of cells of other organism(GO:0051712) |
| 0.6 |
4.4 |
GO:0015755 |
fructose transport(GO:0015755) |
| 0.5 |
2.2 |
GO:0032929 |
negative regulation of superoxide anion generation(GO:0032929) |
| 0.5 |
1.4 |
GO:0060318 |
definitive erythrocyte differentiation(GO:0060318) |
| 0.4 |
1.3 |
GO:0001845 |
phagolysosome assembly(GO:0001845) phagosome maturation(GO:0090382) |
| 0.4 |
1.8 |
GO:1904037 |
positive regulation of epithelial cell apoptotic process(GO:1904037) positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.4 |
2.3 |
GO:0002313 |
mature B cell differentiation involved in immune response(GO:0002313) |
| 0.4 |
2.2 |
GO:0030186 |
melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.4 |
2.2 |
GO:0030277 |
epithelial structure maintenance(GO:0010669) maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.4 |
1.4 |
GO:0071364 |
cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.3 |
2.4 |
GO:0045007 |
depurination(GO:0045007) |
| 0.3 |
1.4 |
GO:0051534 |
negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.3 |
0.7 |
GO:0006307 |
DNA dealkylation involved in DNA repair(GO:0006307) DNA dealkylation(GO:0035510) |
| 0.3 |
2.7 |
GO:0045084 |
positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.3 |
2.7 |
GO:0007064 |
mitotic sister chromatid cohesion(GO:0007064) kinetochore assembly(GO:0051382) kinetochore organization(GO:0051383) |
| 0.3 |
1.0 |
GO:0002538 |
arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.3 |
1.0 |
GO:0002431 |
Fc receptor mediated stimulatory signaling pathway(GO:0002431) regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.3 |
1.0 |
GO:0061298 |
retina vasculature development in camera-type eye(GO:0061298) retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.3 |
7.1 |
GO:0002504 |
antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.3 |
2.8 |
GO:0045078 |
positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.3 |
0.9 |
GO:0016476 |
regulation of embryonic cell shape(GO:0016476) |
| 0.3 |
7.5 |
GO:0050732 |
negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) |
| 0.3 |
0.9 |
GO:0007538 |
primary sex determination(GO:0007538) |
| 0.3 |
1.2 |
GO:0001969 |
activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) positive regulation of activation of membrane attack complex(GO:0001970) positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.3 |
1.2 |
GO:0046967 |
cytosol to ER transport(GO:0046967) |
| 0.3 |
2.0 |
GO:0070779 |
D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.3 |
1.7 |
GO:0010832 |
negative regulation of myotube differentiation(GO:0010832) |
| 0.3 |
1.7 |
GO:0007500 |
mesodermal cell fate determination(GO:0007500) |
| 0.3 |
1.9 |
GO:0031639 |
plasminogen activation(GO:0031639) |
| 0.3 |
2.2 |
GO:0071353 |
cellular response to interleukin-4(GO:0071353) |
| 0.3 |
0.5 |
GO:0030223 |
neutrophil differentiation(GO:0030223) |
| 0.3 |
0.8 |
GO:0060316 |
positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.3 |
2.6 |
GO:0031017 |
exocrine pancreas development(GO:0031017) |
| 0.2 |
1.4 |
GO:0099515 |
nuclear migration along microfilament(GO:0031022) actin filament-based transport(GO:0099515) |
| 0.2 |
1.4 |
GO:0002726 |
positive regulation of T cell cytokine production(GO:0002726) |
| 0.2 |
0.9 |
GO:0001705 |
ectoderm formation(GO:0001705) central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.2 |
0.7 |
GO:0033632 |
regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.2 |
0.9 |
GO:0015868 |
purine ribonucleotide transport(GO:0015868) |
| 0.2 |
0.6 |
GO:0070171 |
negative regulation of tooth mineralization(GO:0070171) |
| 0.2 |
2.1 |
GO:0034372 |
triglyceride-rich lipoprotein particle remodeling(GO:0034370) very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.2 |
0.2 |
GO:0014874 |
response to stimulus involved in regulation of muscle adaptation(GO:0014874) |
| 0.2 |
0.6 |
GO:0019482 |
beta-alanine metabolic process(GO:0019482) |
| 0.2 |
0.4 |
GO:0071380 |
cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.2 |
0.8 |
GO:0033147 |
negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.2 |
1.2 |
GO:0060397 |
JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.2 |
1.2 |
GO:0033160 |
positive regulation of protein import into nucleus, translocation(GO:0033160) |
| 0.2 |
0.2 |
GO:0035021 |
negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.2 |
0.9 |
GO:0060292 |
response to hydroperoxide(GO:0033194) long term synaptic depression(GO:0060292) |
| 0.2 |
3.7 |
GO:0006957 |
complement activation, alternative pathway(GO:0006957) |
| 0.2 |
1.8 |
GO:0045351 |
type I interferon biosynthetic process(GO:0045351) |
| 0.2 |
1.6 |
GO:0070327 |
thyroid hormone transport(GO:0070327) |
| 0.2 |
0.5 |
GO:0010836 |
negative regulation of protein ADP-ribosylation(GO:0010836) negative regulation of protein glycosylation(GO:0060051) |
| 0.2 |
3.0 |
GO:0006071 |
glycerol metabolic process(GO:0006071) |
| 0.2 |
0.5 |
GO:0033578 |
protein glycosylation in Golgi(GO:0033578) |
| 0.2 |
0.7 |
GO:0034382 |
chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) |
| 0.2 |
0.5 |
GO:0043095 |
regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.2 |
0.8 |
GO:0051852 |
disruption by host of symbiont cells(GO:0051852) killing by host of symbiont cells(GO:0051873) |
| 0.2 |
0.2 |
GO:0051902 |
negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.2 |
0.8 |
GO:0018106 |
peptidyl-histidine phosphorylation(GO:0018106) |
| 0.2 |
2.7 |
GO:0050930 |
induction of positive chemotaxis(GO:0050930) |
| 0.2 |
0.8 |
GO:0010759 |
regulation of macrophage chemotaxis(GO:0010758) positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.2 |
0.5 |
GO:0003011 |
diaphragm contraction(GO:0002086) involuntary skeletal muscle contraction(GO:0003011) |
| 0.2 |
1.7 |
GO:0001782 |
B cell homeostasis(GO:0001782) |
| 0.2 |
0.5 |
GO:0046520 |
diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) sphingoid biosynthetic process(GO:0046520) |
| 0.2 |
0.5 |
GO:0043366 |
beta selection(GO:0043366) |
| 0.2 |
0.5 |
GO:0048143 |
astrocyte activation(GO:0048143) |
| 0.1 |
0.9 |
GO:0021978 |
telencephalon regionalization(GO:0021978) |
| 0.1 |
1.0 |
GO:0019264 |
glycine biosynthetic process from serine(GO:0019264) |
| 0.1 |
0.6 |
GO:0000447 |
endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) cleavage involved in rRNA processing(GO:0000469) endonucleolytic cleavage involved in rRNA processing(GO:0000478) endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) RNA 5'-end processing(GO:0000966) RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.1 |
0.9 |
GO:0060710 |
chorio-allantoic fusion(GO:0060710) |
| 0.1 |
0.6 |
GO:0010727 |
negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.1 |
0.6 |
GO:0046778 |
modification by virus of host mRNA processing(GO:0046778) |
| 0.1 |
0.3 |
GO:0071218 |
cellular response to misfolded protein(GO:0071218) |
| 0.1 |
0.7 |
GO:0035455 |
response to interferon-alpha(GO:0035455) |
| 0.1 |
0.4 |
GO:0045198 |
establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.1 |
1.1 |
GO:0007598 |
blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 |
1.1 |
GO:0006670 |
sphingosine metabolic process(GO:0006670) diol metabolic process(GO:0034311) |
| 0.1 |
0.5 |
GO:0046855 |
phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 |
0.9 |
GO:0006572 |
tyrosine catabolic process(GO:0006572) |
| 0.1 |
0.4 |
GO:0090073 |
positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 |
0.5 |
GO:0043922 |
negative regulation by host of viral transcription(GO:0043922) |
| 0.1 |
1.4 |
GO:0007130 |
synaptonemal complex assembly(GO:0007130) |
| 0.1 |
3.8 |
GO:0050853 |
B cell receptor signaling pathway(GO:0050853) |
| 0.1 |
1.2 |
GO:0002920 |
regulation of humoral immune response(GO:0002920) |
| 0.1 |
0.3 |
GO:0033629 |
negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.1 |
0.3 |
GO:0015919 |
peroxisomal membrane transport(GO:0015919) |
| 0.1 |
0.6 |
GO:0050957 |
equilibrioception(GO:0050957) |
| 0.1 |
0.4 |
GO:0007285 |
primary spermatocyte growth(GO:0007285) |
| 0.1 |
0.5 |
GO:0006682 |
galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 |
1.8 |
GO:0030212 |
hyaluronan metabolic process(GO:0030212) |
| 0.1 |
0.4 |
GO:0018916 |
nitrobenzene metabolic process(GO:0018916) |
| 0.1 |
0.1 |
GO:1903519 |
apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.1 |
0.9 |
GO:0030644 |
cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.1 |
0.4 |
GO:0071428 |
rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
| 0.1 |
3.2 |
GO:0050830 |
defense response to Gram-positive bacterium(GO:0050830) |
| 0.1 |
1.2 |
GO:0046653 |
tetrahydrofolate metabolic process(GO:0046653) |
| 0.1 |
0.9 |
GO:0006561 |
proline biosynthetic process(GO:0006561) |
| 0.1 |
0.1 |
GO:0042760 |
very long-chain fatty acid catabolic process(GO:0042760) |
| 0.1 |
0.4 |
GO:0032119 |
sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 |
1.1 |
GO:0061756 |
leukocyte tethering or rolling(GO:0050901) leukocyte adhesion to vascular endothelial cell(GO:0061756) |
| 0.1 |
0.5 |
GO:0050653 |
UDP-N-acetylgalactosamine metabolic process(GO:0019276) chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.1 |
0.5 |
GO:0045299 |
otolith mineralization(GO:0045299) |
| 0.1 |
0.5 |
GO:0021631 |
optic nerve morphogenesis(GO:0021631) |
| 0.1 |
0.5 |
GO:0042816 |
pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.1 |
0.6 |
GO:0010957 |
negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.1 |
0.4 |
GO:0033622 |
integrin activation(GO:0033622) regulation of integrin activation(GO:0033623) positive regulation of integrin activation(GO:0033625) |
| 0.1 |
0.3 |
GO:0006655 |
phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin metabolic process(GO:0032048) cardiolipin biosynthetic process(GO:0032049) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.1 |
0.2 |
GO:0052556 |
pathogen-associated molecular pattern dependent induction by symbiont of host innate immune response(GO:0052033) positive regulation by symbiont of host innate immune response(GO:0052166) modulation by symbiont of host innate immune response(GO:0052167) pathogen-associated molecular pattern dependent modulation by symbiont of host innate immune response(GO:0052169) pathogen-associated molecular pattern dependent induction by organism of innate immune response of other organism involved in symbiotic interaction(GO:0052257) positive regulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052305) modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052306) pathogen-associated molecular pattern dependent modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052308) positive regulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052555) positive regulation by symbiont of host immune response(GO:0052556) |
| 0.1 |
0.4 |
GO:0051124 |
synaptic growth at neuromuscular junction(GO:0051124) |
| 0.1 |
0.7 |
GO:0016557 |
peroxisome membrane biogenesis(GO:0016557) |
| 0.1 |
1.0 |
GO:0046784 |
viral mRNA export from host cell nucleus(GO:0046784) |
| 0.1 |
0.4 |
GO:0048619 |
embryonic hindgut morphogenesis(GO:0048619) |
| 0.1 |
1.6 |
GO:0006750 |
glutathione biosynthetic process(GO:0006750) |
| 0.1 |
4.4 |
GO:0006958 |
complement activation, classical pathway(GO:0006958) |
| 0.1 |
2.5 |
GO:0055074 |
cellular calcium ion homeostasis(GO:0006874) calcium ion homeostasis(GO:0055074) cellular divalent inorganic cation homeostasis(GO:0072503) divalent inorganic cation homeostasis(GO:0072507) |
| 0.1 |
3.4 |
GO:0006749 |
glutathione metabolic process(GO:0006749) |
| 0.1 |
0.9 |
GO:0016998 |
cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 |
0.7 |
GO:0032776 |
DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.1 |
0.9 |
GO:0016199 |
axon midline choice point recognition(GO:0016199) |
| 0.1 |
0.7 |
GO:0033235 |
positive regulation of protein sumoylation(GO:0033235) |
| 0.1 |
1.3 |
GO:0007076 |
mitotic chromosome condensation(GO:0007076) |
| 0.1 |
0.3 |
GO:0010875 |
positive regulation of cholesterol efflux(GO:0010875) |
| 0.1 |
0.4 |
GO:0007281 |
germ cell development(GO:0007281) |
| 0.1 |
0.6 |
GO:0040032 |
post-embryonic body morphogenesis(GO:0040032) |
| 0.1 |
0.6 |
GO:0009649 |
entrainment of circadian clock(GO:0009649) |
| 0.1 |
0.8 |
GO:0048712 |
negative regulation of astrocyte differentiation(GO:0048712) |
| 0.1 |
0.4 |
GO:0045945 |
positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.1 |
1.8 |
GO:0007158 |
neuron cell-cell adhesion(GO:0007158) |
| 0.1 |
0.3 |
GO:0003057 |
regulation of the force of heart contraction by chemical signal(GO:0003057) |
| 0.1 |
0.3 |
GO:0043550 |
regulation of lipid kinase activity(GO:0043550) |
| 0.1 |
7.6 |
GO:0008360 |
regulation of cell shape(GO:0008360) |
| 0.1 |
0.2 |
GO:0030812 |
negative regulation of nucleotide catabolic process(GO:0030812) |
| 0.1 |
0.8 |
GO:0015937 |
coenzyme A biosynthetic process(GO:0015937) |
| 0.1 |
0.3 |
GO:0019805 |
quinolinate biosynthetic process(GO:0019805) |
| 0.1 |
0.2 |
GO:0046208 |
spermine catabolic process(GO:0046208) |
| 0.1 |
0.3 |
GO:0034204 |
lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.1 |
0.7 |
GO:0015886 |
heme transport(GO:0015886) |
| 0.1 |
0.2 |
GO:0001552 |
ovarian follicle atresia(GO:0001552) |
| 0.1 |
0.2 |
GO:0006420 |
arginyl-tRNA aminoacylation(GO:0006420) |
| 0.1 |
0.2 |
GO:0010803 |
regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 0.1 |
1.4 |
GO:0007342 |
fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.1 |
0.4 |
GO:0009439 |
cyanate metabolic process(GO:0009439) cyanate catabolic process(GO:0009440) |
| 0.1 |
0.2 |
GO:0010046 |
response to mycotoxin(GO:0010046) |
| 0.1 |
0.4 |
GO:0003070 |
age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) |
| 0.1 |
5.1 |
GO:0031668 |
cellular response to extracellular stimulus(GO:0031668) |
| 0.1 |
0.5 |
GO:0018027 |
peptidyl-lysine dimethylation(GO:0018027) |
| 0.1 |
1.3 |
GO:0010745 |
negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.1 |
2.1 |
GO:0050918 |
positive chemotaxis(GO:0050918) |
| 0.1 |
0.2 |
GO:0061188 |
regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.1 |
0.6 |
GO:0034331 |
cell junction maintenance(GO:0034331) cell-cell junction maintenance(GO:0045217) |
| 0.1 |
0.2 |
GO:0010834 |
obsolete telomere maintenance via telomere shortening(GO:0010834) |
| 0.1 |
0.4 |
GO:0060259 |
regulation of feeding behavior(GO:0060259) |
| 0.1 |
0.8 |
GO:0030219 |
megakaryocyte differentiation(GO:0030219) |
| 0.1 |
0.1 |
GO:0017183 |
peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 |
0.4 |
GO:0000183 |
chromatin silencing at rDNA(GO:0000183) |
| 0.1 |
0.4 |
GO:0030382 |
sperm mitochondrion organization(GO:0030382) |
| 0.1 |
1.6 |
GO:0051923 |
sulfation(GO:0051923) |
| 0.1 |
0.3 |
GO:0070309 |
lens fiber cell morphogenesis(GO:0070309) |
| 0.1 |
0.3 |
GO:0006436 |
tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 |
0.7 |
GO:0030147 |
obsolete natriuresis(GO:0030147) |
| 0.1 |
2.2 |
GO:0006309 |
DNA catabolic process, endonucleolytic(GO:0000737) apoptotic DNA fragmentation(GO:0006309) |
| 0.1 |
0.2 |
GO:0019919 |
pathogenesis(GO:0009405) peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.1 |
1.1 |
GO:0018298 |
protein-chromophore linkage(GO:0018298) |
| 0.1 |
1.7 |
GO:0006730 |
one-carbon metabolic process(GO:0006730) |
| 0.1 |
0.4 |
GO:0010922 |
positive regulation of phosphatase activity(GO:0010922) positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.1 |
2.2 |
GO:0008206 |
bile acid metabolic process(GO:0008206) |
| 0.1 |
2.3 |
GO:0070830 |
bicellular tight junction assembly(GO:0070830) |
| 0.1 |
3.4 |
GO:0006959 |
humoral immune response(GO:0006959) |
| 0.1 |
1.3 |
GO:0033540 |
fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 |
0.2 |
GO:0001706 |
endoderm formation(GO:0001706) |
| 0.1 |
0.4 |
GO:0006883 |
cellular sodium ion homeostasis(GO:0006883) |
| 0.1 |
0.2 |
GO:0018283 |
metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.1 |
0.2 |
GO:0032959 |
inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.1 |
0.2 |
GO:0006196 |
AMP catabolic process(GO:0006196) |
| 0.1 |
0.6 |
GO:0006198 |
cAMP catabolic process(GO:0006198) |
| 0.1 |
1.6 |
GO:0050728 |
negative regulation of inflammatory response(GO:0050728) |
| 0.1 |
0.3 |
GO:0007144 |
female meiosis I(GO:0007144) |
| 0.1 |
1.0 |
GO:0000226 |
microtubule cytoskeleton organization(GO:0000226) |
| 0.1 |
0.3 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 |
0.3 |
GO:0006422 |
aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.1 |
0.3 |
GO:0032754 |
positive regulation of interleukin-13 production(GO:0032736) positive regulation of interleukin-5 production(GO:0032754) |
| 0.1 |
0.2 |
GO:0060014 |
granulosa cell differentiation(GO:0060014) |
| 0.1 |
0.3 |
GO:0030951 |
establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 |
0.3 |
GO:0006975 |
DNA damage induced protein phosphorylation(GO:0006975) |
| 0.1 |
0.9 |
GO:0045885 |
obsolete positive regulation of survival gene product expression(GO:0045885) |
| 0.1 |
0.5 |
GO:0090080 |
positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.1 |
0.1 |
GO:0033344 |
cholesterol efflux(GO:0033344) |
| 0.1 |
0.2 |
GO:0032074 |
negative regulation of nuclease activity(GO:0032074) |
| 0.1 |
0.3 |
GO:0033006 |
regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.1 |
0.5 |
GO:0031116 |
positive regulation of microtubule polymerization(GO:0031116) |
| 0.1 |
0.5 |
GO:0022027 |
interkinetic nuclear migration(GO:0022027) |
| 0.1 |
0.2 |
GO:0010761 |
fibroblast migration(GO:0010761) |
| 0.1 |
1.0 |
GO:0007032 |
endosome organization(GO:0007032) |
| 0.1 |
0.6 |
GO:0071347 |
cellular response to interleukin-1(GO:0071347) |
| 0.1 |
1.3 |
GO:0015804 |
neutral amino acid transport(GO:0015804) |
| 0.0 |
0.1 |
GO:0045750 |
obsolete positive regulation of S phase of mitotic cell cycle(GO:0045750) |
| 0.0 |
0.1 |
GO:0035414 |
negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 |
0.6 |
GO:0090399 |
replicative senescence(GO:0090399) |
| 0.0 |
0.3 |
GO:0019227 |
neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 |
0.6 |
GO:1903078 |
positive regulation of establishment of protein localization to plasma membrane(GO:0090004) positive regulation of protein localization to plasma membrane(GO:1903078) positive regulation of protein localization to cell periphery(GO:1904377) |
| 0.0 |
0.8 |
GO:0000303 |
response to superoxide(GO:0000303) |
| 0.0 |
0.8 |
GO:0042438 |
melanin metabolic process(GO:0006582) melanin biosynthetic process(GO:0042438) |
| 0.0 |
0.8 |
GO:0015893 |
drug transport(GO:0015893) |
| 0.0 |
1.7 |
GO:0000289 |
nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 |
0.4 |
GO:0006013 |
mannose metabolic process(GO:0006013) |
| 0.0 |
0.1 |
GO:0060122 |
auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 |
2.0 |
GO:0043488 |
regulation of RNA stability(GO:0043487) regulation of mRNA stability(GO:0043488) |
| 0.0 |
0.1 |
GO:0032223 |
negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) neurotransmitter receptor metabolic process(GO:0045213) |
| 0.0 |
0.2 |
GO:0002092 |
positive regulation of receptor internalization(GO:0002092) |
| 0.0 |
0.4 |
GO:0016180 |
snRNA processing(GO:0016180) |
| 0.0 |
0.4 |
GO:0050848 |
regulation of calcium-mediated signaling(GO:0050848) |
| 0.0 |
0.2 |
GO:0021988 |
olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 |
0.2 |
GO:0021615 |
glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 |
0.1 |
GO:0051967 |
negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 |
0.3 |
GO:0016561 |
protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.0 |
0.1 |
GO:0072385 |
minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 |
0.3 |
GO:0008272 |
sulfate transport(GO:0008272) |
| 0.0 |
3.3 |
GO:0042113 |
B cell activation(GO:0042113) |
| 0.0 |
0.3 |
GO:0043545 |
Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 |
0.7 |
GO:0006596 |
polyamine biosynthetic process(GO:0006596) |
| 0.0 |
0.9 |
GO:0008211 |
glucocorticoid metabolic process(GO:0008211) |
| 0.0 |
0.4 |
GO:0045793 |
positive regulation of cell size(GO:0045793) |
| 0.0 |
1.9 |
GO:0007202 |
activation of phospholipase C activity(GO:0007202) |
| 0.0 |
0.8 |
GO:0042098 |
T cell proliferation(GO:0042098) |
| 0.0 |
0.5 |
GO:0043252 |
sodium-independent organic anion transport(GO:0043252) |
| 0.0 |
1.1 |
GO:0034199 |
activation of protein kinase A activity(GO:0034199) |
| 0.0 |
1.1 |
GO:0051592 |
response to calcium ion(GO:0051592) |
| 0.0 |
0.2 |
GO:0090178 |
outflow tract septum morphogenesis(GO:0003148) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 |
0.2 |
GO:0030643 |
cellular phosphate ion homeostasis(GO:0030643) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 |
0.2 |
GO:0032020 |
ISG15-protein conjugation(GO:0032020) |
| 0.0 |
0.2 |
GO:2000171 |
negative regulation of dendrite development(GO:2000171) |
| 0.0 |
3.8 |
GO:0008203 |
cholesterol metabolic process(GO:0008203) |
| 0.0 |
0.0 |
GO:0046022 |
regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.0 |
0.3 |
GO:0060056 |
mammary gland involution(GO:0060056) |
| 0.0 |
0.2 |
GO:0050665 |
hydrogen peroxide biosynthetic process(GO:0050665) |
| 0.0 |
0.3 |
GO:2000108 |
regulation of protein homooligomerization(GO:0032462) positive regulation of protein homooligomerization(GO:0032464) positive regulation of lymphocyte apoptotic process(GO:0070230) positive regulation of T cell apoptotic process(GO:0070234) positive regulation of thymocyte apoptotic process(GO:0070245) positive regulation of leukocyte apoptotic process(GO:2000108) |
| 0.0 |
0.7 |
GO:0000045 |
autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.0 |
0.8 |
GO:0016254 |
preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 |
0.3 |
GO:0007351 |
tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.0 |
0.2 |
GO:0046655 |
folic acid metabolic process(GO:0046655) |
| 0.0 |
4.0 |
GO:0050852 |
T cell receptor signaling pathway(GO:0050852) |
| 0.0 |
0.2 |
GO:0008343 |
adult feeding behavior(GO:0008343) |
| 0.0 |
0.2 |
GO:0016322 |
neuron remodeling(GO:0016322) |
| 0.0 |
0.3 |
GO:0015695 |
organic cation transport(GO:0015695) |
| 0.0 |
2.0 |
GO:0007623 |
circadian rhythm(GO:0007623) |
| 0.0 |
0.3 |
GO:0001510 |
RNA methylation(GO:0001510) |
| 0.0 |
0.3 |
GO:0071445 |
obsolete cellular response to protein stimulus(GO:0071445) |
| 0.0 |
0.7 |
GO:0001938 |
positive regulation of endothelial cell proliferation(GO:0001938) |
| 0.0 |
1.3 |
GO:0055088 |
lipid homeostasis(GO:0055088) |
| 0.0 |
0.1 |
GO:0048934 |
peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 |
0.7 |
GO:0006084 |
acetyl-CoA metabolic process(GO:0006084) |
| 0.0 |
0.3 |
GO:0000042 |
protein targeting to Golgi(GO:0000042) establishment of protein localization to Golgi(GO:0072600) |
| 0.0 |
0.1 |
GO:0048251 |
elastic fiber assembly(GO:0048251) |
| 0.0 |
0.2 |
GO:0051491 |
positive regulation of filopodium assembly(GO:0051491) |
| 0.0 |
0.3 |
GO:0006298 |
mismatch repair(GO:0006298) |
| 0.0 |
0.2 |
GO:0042637 |
catagen(GO:0042637) |
| 0.0 |
0.1 |
GO:0071168 |
negative regulation of sister chromatid cohesion(GO:0045875) protein localization to chromatin(GO:0071168) |
| 0.0 |
0.2 |
GO:0019471 |
peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 |
0.2 |
GO:0060155 |
platelet dense granule organization(GO:0060155) |
| 0.0 |
0.2 |
GO:0060391 |
positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 |
0.2 |
GO:0010842 |
retina layer formation(GO:0010842) |
| 0.0 |
0.4 |
GO:0035036 |
binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) |
| 0.0 |
0.2 |
GO:0006398 |
mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 |
0.7 |
GO:0051925 |
obsolete regulation of calcium ion transport via voltage-gated calcium channel activity(GO:0051925) |
| 0.0 |
0.3 |
GO:0048048 |
embryonic eye morphogenesis(GO:0048048) |
| 0.0 |
2.6 |
GO:0035023 |
regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 |
0.1 |
GO:0043029 |
T cell homeostasis(GO:0043029) |
| 0.0 |
0.1 |
GO:0009992 |
cellular water homeostasis(GO:0009992) |
| 0.0 |
2.1 |
GO:0071357 |
response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 |
0.3 |
GO:0006706 |
steroid catabolic process(GO:0006706) |
| 0.0 |
0.2 |
GO:0051290 |
protein heterotetramerization(GO:0051290) |
| 0.0 |
0.1 |
GO:0006930 |
substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 |
0.5 |
GO:0006386 |
transcription elongation from RNA polymerase III promoter(GO:0006385) termination of RNA polymerase III transcription(GO:0006386) |
| 0.0 |
0.2 |
GO:0006359 |
regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 |
0.2 |
GO:0010629 |
negative regulation of gene expression(GO:0010629) |
| 0.0 |
0.2 |
GO:0070936 |
protein K48-linked ubiquitination(GO:0070936) |
| 0.0 |
0.1 |
GO:0016246 |
RNA interference(GO:0016246) production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 |
0.1 |
GO:1901503 |
ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 |
0.1 |
GO:0002209 |
behavioral fear response(GO:0001662) behavioral defense response(GO:0002209) |
| 0.0 |
0.5 |
GO:0007157 |
heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 |
0.2 |
GO:0009120 |
deoxyribonucleoside metabolic process(GO:0009120) |
| 0.0 |
0.2 |
GO:0045003 |
DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 |
0.2 |
GO:0030512 |
negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 |
0.1 |
GO:0090400 |
stress-induced premature senescence(GO:0090400) |
| 0.0 |
0.3 |
GO:0001556 |
oocyte maturation(GO:0001556) |
| 0.0 |
0.3 |
GO:0045197 |
establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 |
0.1 |
GO:0048702 |
embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 |
0.2 |
GO:0009650 |
UV protection(GO:0009650) |
| 0.0 |
0.2 |
GO:0051865 |
protein autoubiquitination(GO:0051865) |
| 0.0 |
0.1 |
GO:0060992 |
response to fungicide(GO:0060992) |
| 0.0 |
0.2 |
GO:0051438 |
regulation of ubiquitin-protein transferase activity(GO:0051438) |
| 0.0 |
0.2 |
GO:0006002 |
fructose 6-phosphate metabolic process(GO:0006002) |
| 0.0 |
0.1 |
GO:0090292 |
nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 |
0.6 |
GO:0007602 |
phototransduction(GO:0007602) |
| 0.0 |
0.3 |
GO:0000060 |
protein import into nucleus, translocation(GO:0000060) |
| 0.0 |
0.1 |
GO:0018344 |
protein geranylgeranylation(GO:0018344) |
| 0.0 |
0.2 |
GO:0008535 |
respiratory chain complex IV assembly(GO:0008535) |
| 0.0 |
0.3 |
GO:0034508 |
centromere complex assembly(GO:0034508) |
| 0.0 |
0.1 |
GO:0007549 |
dosage compensation(GO:0007549) |
| 0.0 |
0.6 |
GO:0006024 |
glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.0 |
0.1 |
GO:0030263 |
apoptotic chromosome condensation(GO:0030263) |
| 0.0 |
0.4 |
GO:0045646 |
regulation of erythrocyte differentiation(GO:0045646) |
| 0.0 |
0.2 |
GO:0030520 |
intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.0 |
0.2 |
GO:0006734 |
NADH metabolic process(GO:0006734) |
| 0.0 |
0.1 |
GO:0009301 |
snRNA transcription(GO:0009301) |
| 0.0 |
0.4 |
GO:0010827 |
regulation of glucose transport(GO:0010827) |
| 0.0 |
0.2 |
GO:0019228 |
neuronal action potential(GO:0019228) |
| 0.0 |
0.3 |
GO:0034605 |
cellular response to heat(GO:0034605) |
| 0.0 |
0.2 |
GO:0007194 |
negative regulation of adenylate cyclase activity(GO:0007194) negative regulation of cyclase activity(GO:0031280) negative regulation of lyase activity(GO:0051350) |
| 0.0 |
0.5 |
GO:0015985 |
energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 |
0.1 |
GO:0000491 |
small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 |
0.3 |
GO:0042475 |
odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 |
0.0 |
GO:0097237 |
response to cobalt ion(GO:0032025) cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.0 |
0.3 |
GO:0071577 |
zinc II ion transmembrane transport(GO:0071577) |
| 0.0 |
0.1 |
GO:0032203 |
telomere formation via telomerase(GO:0032203) |
| 0.0 |
0.1 |
GO:0048013 |
ephrin receptor signaling pathway(GO:0048013) |
| 0.0 |
0.3 |
GO:0006541 |
glutamine metabolic process(GO:0006541) |
| 0.0 |
0.0 |
GO:0030432 |
peristalsis(GO:0030432) |
| 0.0 |
0.0 |
GO:0003351 |
epithelial cilium movement(GO:0003351) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 |
0.4 |
GO:0006401 |
RNA catabolic process(GO:0006401) |
| 0.0 |
0.1 |
GO:0006760 |
folic acid-containing compound metabolic process(GO:0006760) |
| 0.0 |
2.0 |
GO:0042254 |
ribosome biogenesis(GO:0042254) |
| 0.0 |
0.1 |
GO:0042136 |
neurotransmitter biosynthetic process(GO:0042136) |
| 0.0 |
0.3 |
GO:0001578 |
microtubule bundle formation(GO:0001578) |
| 0.0 |
0.3 |
GO:0043968 |
histone H2A acetylation(GO:0043968) |
| 0.0 |
0.1 |
GO:0045872 |
positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 |
0.2 |
GO:0043101 |
purine-containing compound salvage(GO:0043101) |
| 0.0 |
0.1 |
GO:0006516 |
glycoprotein catabolic process(GO:0006516) |
| 0.0 |
0.0 |
GO:0021904 |
dorsal/ventral neural tube patterning(GO:0021904) |
| 0.0 |
0.2 |
GO:0046718 |
viral entry into host cell(GO:0046718) |
| 0.0 |
0.2 |
GO:0032720 |
negative regulation of tumor necrosis factor production(GO:0032720) negative regulation of tumor necrosis factor superfamily cytokine production(GO:1903556) |
| 0.0 |
0.1 |
GO:0003333 |
amino acid transmembrane transport(GO:0003333) |
| 0.0 |
0.1 |
GO:0006491 |
N-glycan processing(GO:0006491) |
| 0.0 |
0.1 |
GO:0048477 |
oogenesis(GO:0048477) |
| 0.0 |
0.2 |
GO:0045773 |
positive regulation of axon extension(GO:0045773) |
| 0.0 |
0.1 |
GO:0040034 |
regulation of development, heterochronic(GO:0040034) regulation of timing of cell differentiation(GO:0048505) |
| 0.0 |
0.4 |
GO:0030073 |
insulin secretion(GO:0030073) |
| 0.0 |
0.2 |
GO:0007131 |
reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 |
0.1 |
GO:0070304 |
positive regulation of stress-activated MAPK cascade(GO:0032874) positive regulation of stress-activated protein kinase signaling cascade(GO:0070304) |
| 0.0 |
0.1 |
GO:0015758 |
hexose transport(GO:0008645) monosaccharide transport(GO:0015749) glucose transport(GO:0015758) |
| 0.0 |
0.1 |
GO:0061448 |
connective tissue development(GO:0061448) |
| 0.0 |
0.1 |
GO:0060644 |
mammary gland epithelial cell differentiation(GO:0060644) |
| 0.0 |
0.2 |
GO:0009168 |
ATP biosynthetic process(GO:0006754) purine nucleoside monophosphate biosynthetic process(GO:0009127) nucleoside triphosphate biosynthetic process(GO:0009142) purine nucleoside triphosphate biosynthetic process(GO:0009145) purine ribonucleoside monophosphate biosynthetic process(GO:0009168) ribonucleoside triphosphate biosynthetic process(GO:0009201) purine ribonucleoside triphosphate biosynthetic process(GO:0009206) |
| 0.0 |
0.2 |
GO:0032012 |
regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 |
0.1 |
GO:0030878 |
thyroid gland development(GO:0030878) |
| 0.0 |
0.3 |
GO:0046470 |
phosphatidylcholine metabolic process(GO:0046470) |
| 0.0 |
0.1 |
GO:0060706 |
cell differentiation involved in embryonic placenta development(GO:0060706) |
| 0.0 |
0.3 |
GO:0008543 |
fibroblast growth factor receptor signaling pathway(GO:0008543) cellular response to fibroblast growth factor stimulus(GO:0044344) response to fibroblast growth factor(GO:0071774) |
| 0.0 |
0.4 |
GO:0001764 |
neuron migration(GO:0001764) |
| 0.0 |
0.1 |
GO:0008089 |
anterograde axonal transport(GO:0008089) |