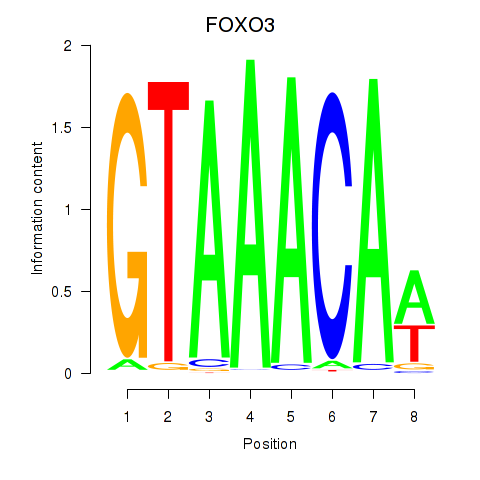
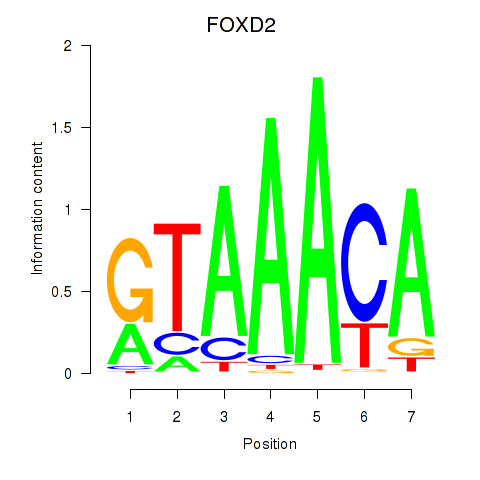
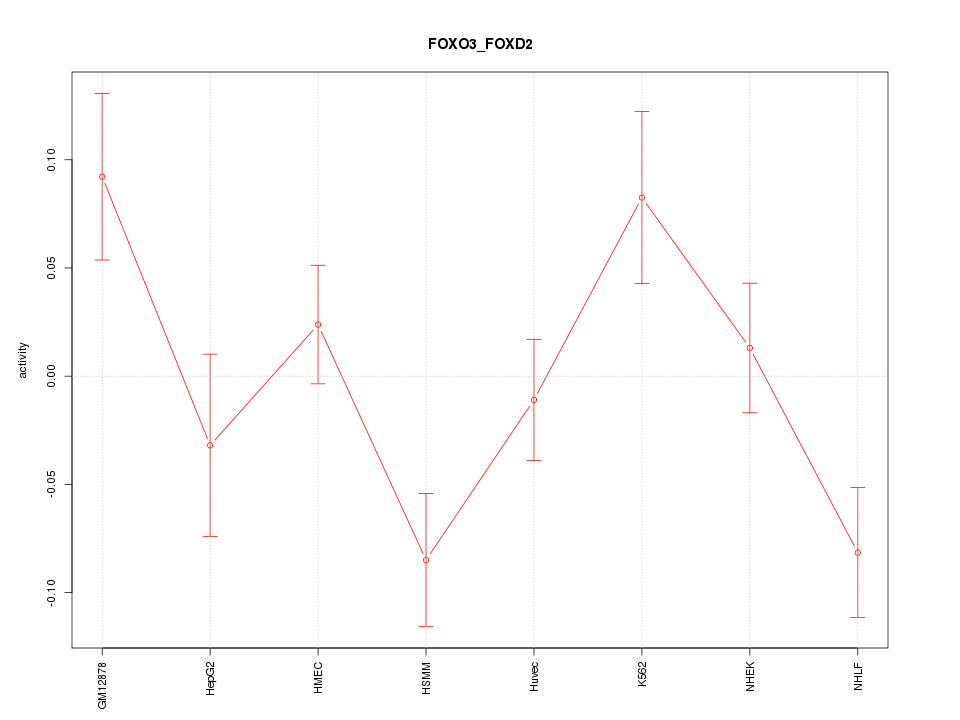
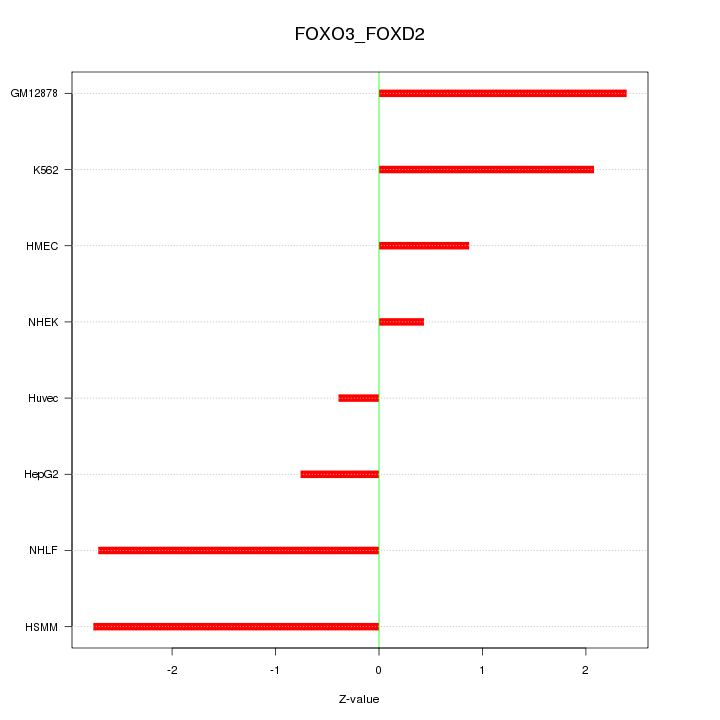
| 1.2 |
4.7 |
GO:0060649 |
nipple development(GO:0060618) mammary gland bud elongation(GO:0060649) nipple morphogenesis(GO:0060658) nipple sheath formation(GO:0060659) |
| 1.0 |
1.0 |
GO:0003006 |
developmental process involved in reproduction(GO:0003006) |
| 0.8 |
2.5 |
GO:1901724 |
positive regulation of cell proliferation involved in kidney development(GO:1901724) |
| 0.7 |
2.6 |
GO:0031580 |
membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.6 |
1.9 |
GO:0045603 |
positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.6 |
2.5 |
GO:0010157 |
response to chlorate(GO:0010157) |
| 0.6 |
1.8 |
GO:0035461 |
vitamin transmembrane transport(GO:0035461) |
| 0.4 |
1.3 |
GO:0071603 |
negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) endothelial cell-cell adhesion(GO:0071603) |
| 0.4 |
2.5 |
GO:0006196 |
AMP catabolic process(GO:0006196) |
| 0.4 |
1.7 |
GO:0042320 |
regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) |
| 0.4 |
1.2 |
GO:0010430 |
ethanol catabolic process(GO:0006068) fatty acid omega-oxidation(GO:0010430) |
| 0.4 |
1.6 |
GO:0051573 |
negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.4 |
1.2 |
GO:0002035 |
brain renin-angiotensin system(GO:0002035) |
| 0.4 |
2.4 |
GO:0007352 |
zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.4 |
0.4 |
GO:0034310 |
primary alcohol catabolic process(GO:0034310) |
| 0.4 |
1.4 |
GO:0045338 |
farnesyl diphosphate metabolic process(GO:0045338) |
| 0.4 |
1.4 |
GO:0019805 |
quinolinate biosynthetic process(GO:0019805) |
| 0.4 |
1.8 |
GO:0030951 |
establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.3 |
1.4 |
GO:0021759 |
globus pallidus development(GO:0021759) menarche(GO:0042696) |
| 0.3 |
1.3 |
GO:0002318 |
myeloid progenitor cell differentiation(GO:0002318) |
| 0.3 |
0.9 |
GO:0002291 |
T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.3 |
1.2 |
GO:0043490 |
malate-aspartate shuttle(GO:0043490) |
| 0.3 |
1.2 |
GO:0045085 |
negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.3 |
2.6 |
GO:1900087 |
traversing start control point of mitotic cell cycle(GO:0007089) positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) positive regulation of cell cycle G1/S phase transition(GO:1902808) |
| 0.3 |
0.9 |
GO:0014900 |
smooth muscle hyperplasia(GO:0014806) muscle hyperplasia(GO:0014900) positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.3 |
1.8 |
GO:0061029 |
eyelid development in camera-type eye(GO:0061029) |
| 0.3 |
0.8 |
GO:2000078 |
epithelial cilium movement(GO:0003351) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) regulation of type B pancreatic cell development(GO:2000074) positive regulation of type B pancreatic cell development(GO:2000078) |
| 0.3 |
1.5 |
GO:0006883 |
cellular sodium ion homeostasis(GO:0006883) |
| 0.2 |
0.7 |
GO:0001780 |
neutrophil homeostasis(GO:0001780) |
| 0.2 |
0.7 |
GO:0060120 |
auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.2 |
1.4 |
GO:0031665 |
negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.2 |
10.4 |
GO:0002286 |
T cell activation involved in immune response(GO:0002286) |
| 0.2 |
4.1 |
GO:0016601 |
Rac protein signal transduction(GO:0016601) |
| 0.2 |
1.1 |
GO:0043402 |
glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.2 |
3.5 |
GO:0007020 |
microtubule nucleation(GO:0007020) |
| 0.2 |
0.7 |
GO:0072203 |
kidney mesenchymal cell proliferation(GO:0072135) metanephric mesenchymal cell proliferation involved in metanephros development(GO:0072136) cell proliferation involved in metanephros development(GO:0072203) |
| 0.2 |
1.7 |
GO:0045039 |
protein import into mitochondrial inner membrane(GO:0045039) |
| 0.2 |
0.9 |
GO:0043922 |
negative regulation by host of viral transcription(GO:0043922) |
| 0.2 |
0.9 |
GO:0070527 |
integrin activation(GO:0033622) regulation of integrin activation(GO:0033623) positive regulation of integrin activation(GO:0033625) platelet aggregation(GO:0070527) |
| 0.2 |
0.4 |
GO:0045074 |
negative regulation of interleukin-10 production(GO:0032693) interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.2 |
1.0 |
GO:0017055 |
negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.2 |
0.6 |
GO:0045759 |
negative regulation of action potential(GO:0045759) |
| 0.2 |
0.6 |
GO:0048852 |
diencephalon morphogenesis(GO:0048852) |
| 0.2 |
0.8 |
GO:0032817 |
regulation of natural killer cell proliferation(GO:0032817) |
| 0.2 |
2.1 |
GO:0001706 |
endoderm formation(GO:0001706) |
| 0.2 |
0.6 |
GO:0046521 |
sphingoid catabolic process(GO:0046521) |
| 0.2 |
0.5 |
GO:0070171 |
negative regulation of histone methylation(GO:0031061) negative regulation of tooth mineralization(GO:0070171) |
| 0.2 |
0.3 |
GO:0008050 |
female courtship behavior(GO:0008050) |
| 0.2 |
3.5 |
GO:0007158 |
neuron cell-cell adhesion(GO:0007158) |
| 0.2 |
0.6 |
GO:1904037 |
positive regulation of epithelial cell apoptotic process(GO:1904037) positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.2 |
2.2 |
GO:0045070 |
positive regulation of viral genome replication(GO:0045070) |
| 0.2 |
0.6 |
GO:0046022 |
regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.2 |
0.8 |
GO:0071542 |
dopaminergic neuron differentiation(GO:0071542) |
| 0.2 |
0.8 |
GO:0010508 |
positive regulation of autophagy(GO:0010508) |
| 0.2 |
1.2 |
GO:0042149 |
cellular response to glucose starvation(GO:0042149) |
| 0.1 |
0.9 |
GO:0032825 |
regulation of natural killer cell differentiation(GO:0032823) positive regulation of natural killer cell differentiation(GO:0032825) |
| 0.1 |
1.0 |
GO:0015671 |
oxygen transport(GO:0015671) |
| 0.1 |
0.4 |
GO:0033364 |
mast cell secretory granule organization(GO:0033364) |
| 0.1 |
5.4 |
GO:0006271 |
DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.1 |
0.4 |
GO:0031034 |
skeletal muscle myosin thick filament assembly(GO:0030241) myosin filament assembly(GO:0031034) striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 |
0.4 |
GO:0007518 |
myoblast fate determination(GO:0007518) |
| 0.1 |
2.5 |
GO:1904031 |
positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.1 |
3.7 |
GO:0033209 |
tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.1 |
0.5 |
GO:0030259 |
lipid glycosylation(GO:0030259) |
| 0.1 |
0.4 |
GO:0006288 |
base-excision repair, DNA ligation(GO:0006288) |
| 0.1 |
0.3 |
GO:0050862 |
positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.1 |
0.4 |
GO:1904396 |
regulation of synaptic growth at neuromuscular junction(GO:0008582) regulation of neuromuscular junction development(GO:1904396) |
| 0.1 |
0.4 |
GO:0035623 |
renal glucose absorption(GO:0035623) |
| 0.1 |
0.5 |
GO:0045589 |
regulation of regulatory T cell differentiation(GO:0045589) |
| 0.1 |
2.1 |
GO:0006906 |
vesicle fusion(GO:0006906) |
| 0.1 |
0.5 |
GO:0002052 |
positive regulation of neuroblast proliferation(GO:0002052) |
| 0.1 |
2.0 |
GO:0050930 |
induction of positive chemotaxis(GO:0050930) |
| 0.1 |
0.6 |
GO:0031936 |
negative regulation of chromatin silencing(GO:0031936) |
| 0.1 |
0.6 |
GO:0018106 |
peptidyl-histidine phosphorylation(GO:0018106) |
| 0.1 |
0.1 |
GO:0032656 |
interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) negative regulation of interleukin-13 production(GO:0032696) negative regulation of interleukin-5 production(GO:0032714) |
| 0.1 |
0.4 |
GO:0006391 |
transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 |
0.4 |
GO:0071353 |
cellular response to interleukin-4(GO:0071353) |
| 0.1 |
0.7 |
GO:0048853 |
forebrain morphogenesis(GO:0048853) |
| 0.1 |
0.2 |
GO:0051151 |
negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 |
0.3 |
GO:0071104 |
response to interleukin-9(GO:0071104) response to interleukin-11(GO:0071105) |
| 0.1 |
1.3 |
GO:0032876 |
regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.1 |
0.3 |
GO:0060267 |
transformation of host cell by virus(GO:0019087) positive regulation of respiratory burst(GO:0060267) |
| 0.1 |
0.7 |
GO:0006177 |
GMP biosynthetic process(GO:0006177) |
| 0.1 |
0.3 |
GO:0048865 |
stem cell fate commitment(GO:0048865) stem cell fate determination(GO:0048867) |
| 0.1 |
0.8 |
GO:0048535 |
positive regulation of interleukin-12 biosynthetic process(GO:0045084) lymph node development(GO:0048535) |
| 0.1 |
0.3 |
GO:0032416 |
negative regulation of sodium:proton antiporter activity(GO:0032416) positive regulation of dopamine receptor signaling pathway(GO:0060161) negative regulation of sodium ion transmembrane transport(GO:1902306) negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.1 |
1.0 |
GO:0006975 |
DNA damage induced protein phosphorylation(GO:0006975) |
| 0.1 |
0.4 |
GO:0046621 |
negative regulation of organ growth(GO:0046621) |
| 0.1 |
1.6 |
GO:0045730 |
respiratory burst(GO:0045730) |
| 0.1 |
0.3 |
GO:0006542 |
glutamine biosynthetic process(GO:0006542) |
| 0.1 |
1.6 |
GO:0046854 |
phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 |
0.3 |
GO:0019056 |
modulation by virus of host transcription(GO:0019056) regulation of sprouting of injured axon(GO:0048686) positive regulation of sprouting of injured axon(GO:0048687) regulation of axon extension involved in regeneration(GO:0048690) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.1 |
0.3 |
GO:0040034 |
regulation of development, heterochronic(GO:0040034) regulation of timing of cell differentiation(GO:0048505) |
| 0.1 |
1.3 |
GO:0008625 |
extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) extrinsic apoptotic signaling pathway(GO:0097191) |
| 0.1 |
3.9 |
GO:0070374 |
positive regulation of ERK1 and ERK2 cascade(GO:0070374) |
| 0.1 |
0.3 |
GO:0006531 |
aspartate metabolic process(GO:0006531) aspartate catabolic process(GO:0006533) |
| 0.1 |
0.5 |
GO:0090050 |
positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.1 |
0.9 |
GO:0048585 |
negative regulation of response to stimulus(GO:0048585) |
| 0.1 |
2.0 |
GO:0000291 |
nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.1 |
1.4 |
GO:0006959 |
humoral immune response(GO:0006959) |
| 0.1 |
0.2 |
GO:0034498 |
early endosome to Golgi transport(GO:0034498) |
| 0.1 |
1.1 |
GO:0030889 |
negative regulation of B cell proliferation(GO:0030889) |
| 0.1 |
0.7 |
GO:0001932 |
regulation of protein phosphorylation(GO:0001932) |
| 0.1 |
4.8 |
GO:0030183 |
B cell differentiation(GO:0030183) |
| 0.1 |
0.4 |
GO:0071168 |
protein localization to chromatin(GO:0071168) |
| 0.1 |
0.7 |
GO:0032464 |
positive regulation of protein homooligomerization(GO:0032464) |
| 0.1 |
0.7 |
GO:0045779 |
negative regulation of bone resorption(GO:0045779) |
| 0.1 |
1.2 |
GO:0030260 |
entry into host cell(GO:0030260) entry into host(GO:0044409) entry into cell of other organism involved in symbiotic interaction(GO:0051806) entry into other organism involved in symbiotic interaction(GO:0051828) |
| 0.1 |
0.4 |
GO:0045722 |
positive regulation of gluconeogenesis(GO:0045722) |
| 0.1 |
1.8 |
GO:0006309 |
DNA catabolic process, endonucleolytic(GO:0000737) apoptotic DNA fragmentation(GO:0006309) |
| 0.1 |
0.4 |
GO:0055009 |
atrial cardiac muscle tissue development(GO:0003228) atrial cardiac muscle tissue morphogenesis(GO:0055009) |
| 0.1 |
0.6 |
GO:0043497 |
regulation of protein heterodimerization activity(GO:0043497) |
| 0.1 |
1.6 |
GO:0071577 |
zinc II ion transmembrane transport(GO:0071577) |
| 0.1 |
0.7 |
GO:0046784 |
viral mRNA export from host cell nucleus(GO:0046784) |
| 0.1 |
6.0 |
GO:0031295 |
lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.1 |
0.3 |
GO:0010447 |
response to acidic pH(GO:0010447) |
| 0.1 |
1.1 |
GO:0042730 |
fibrinolysis(GO:0042730) |
| 0.1 |
0.7 |
GO:0007494 |
midgut development(GO:0007494) |
| 0.1 |
0.2 |
GO:0043276 |
anoikis(GO:0043276) |
| 0.1 |
0.5 |
GO:0050912 |
detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) detection of chemical stimulus involved in sensory perception of taste(GO:0050912) |
| 0.1 |
0.6 |
GO:0045616 |
regulation of keratinocyte differentiation(GO:0045616) |
| 0.1 |
0.3 |
GO:0043568 |
positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.1 |
0.9 |
GO:0034440 |
fatty acid oxidation(GO:0019395) lipid oxidation(GO:0034440) |
| 0.1 |
0.5 |
GO:0042572 |
retinol metabolic process(GO:0042572) |
| 0.1 |
0.9 |
GO:0032720 |
negative regulation of tumor necrosis factor production(GO:0032720) negative regulation of tumor necrosis factor superfamily cytokine production(GO:1903556) |
| 0.1 |
0.7 |
GO:0001893 |
maternal placenta development(GO:0001893) |
| 0.1 |
0.5 |
GO:0007141 |
male meiosis I(GO:0007141) |
| 0.1 |
1.8 |
GO:0007214 |
gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 |
0.7 |
GO:0007076 |
mitotic chromosome condensation(GO:0007076) |
| 0.1 |
0.9 |
GO:0034605 |
cellular response to heat(GO:0034605) |
| 0.1 |
2.6 |
GO:0043484 |
regulation of RNA splicing(GO:0043484) |
| 0.0 |
0.7 |
GO:0043330 |
response to exogenous dsRNA(GO:0043330) |
| 0.0 |
0.3 |
GO:0002430 |
complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 |
0.2 |
GO:0030037 |
actin filament reorganization involved in cell cycle(GO:0030037) actin filament reorganization(GO:0090527) |
| 0.0 |
0.3 |
GO:0001660 |
fever generation(GO:0001660) |
| 0.0 |
0.2 |
GO:0032959 |
inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.0 |
1.0 |
GO:0006957 |
complement activation, alternative pathway(GO:0006957) |
| 0.0 |
0.1 |
GO:0042074 |
cell migration involved in gastrulation(GO:0042074) |
| 0.0 |
2.4 |
GO:0006120 |
mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 |
0.3 |
GO:0090151 |
establishment of protein localization to mitochondrial membrane(GO:0090151) |
| 0.0 |
0.3 |
GO:0044245 |
polysaccharide digestion(GO:0044245) |
| 0.0 |
0.1 |
GO:2000831 |
renin-angiotensin regulation of aldosterone production(GO:0002018) regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) positive regulation of NAD(P)H oxidase activity(GO:0033864) corticosteroid hormone secretion(GO:0035930) mineralocorticoid secretion(GO:0035931) aldosterone secretion(GO:0035932) regulation of steroid hormone secretion(GO:2000831) regulation of corticosteroid hormone secretion(GO:2000846) regulation of mineralocorticoid secretion(GO:2000855) regulation of aldosterone secretion(GO:2000858) |
| 0.0 |
0.3 |
GO:0016098 |
monoterpenoid metabolic process(GO:0016098) |
| 0.0 |
0.2 |
GO:0070071 |
response to fructose(GO:0009750) proton-transporting two-sector ATPase complex assembly(GO:0070071) |
| 0.0 |
0.2 |
GO:0070254 |
mucus secretion(GO:0070254) |
| 0.0 |
0.8 |
GO:0051353 |
positive regulation of oxidoreductase activity(GO:0051353) |
| 0.0 |
0.2 |
GO:0007416 |
synapse assembly(GO:0007416) |
| 0.0 |
1.0 |
GO:0006783 |
heme biosynthetic process(GO:0006783) |
| 0.0 |
0.2 |
GO:0008228 |
opsonization(GO:0008228) |
| 0.0 |
0.5 |
GO:0019228 |
neuronal action potential(GO:0019228) |
| 0.0 |
1.9 |
GO:0007193 |
adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 |
0.0 |
GO:0060684 |
epithelial-mesenchymal cell signaling(GO:0060684) |
| 0.0 |
0.5 |
GO:0033138 |
positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 |
0.7 |
GO:0042102 |
positive regulation of T cell proliferation(GO:0042102) |
| 0.0 |
0.4 |
GO:0032438 |
melanosome organization(GO:0032438) pigment granule organization(GO:0048753) |
| 0.0 |
0.1 |
GO:0051823 |
regulation of synapse structural plasticity(GO:0051823) |
| 0.0 |
2.9 |
GO:0007163 |
establishment or maintenance of cell polarity(GO:0007163) |
| 0.0 |
0.8 |
GO:0050870 |
positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.0 |
2.8 |
GO:0097696 |
JAK-STAT cascade(GO:0007259) STAT cascade(GO:0097696) |
| 0.0 |
0.3 |
GO:0060042 |
retina morphogenesis in camera-type eye(GO:0060042) |
| 0.0 |
0.2 |
GO:0006422 |
aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 |
0.2 |
GO:0042797 |
5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 |
0.1 |
GO:0046618 |
drug export(GO:0046618) |
| 0.0 |
1.2 |
GO:0000725 |
double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.0 |
0.3 |
GO:0043537 |
negative regulation of blood vessel endothelial cell migration(GO:0043537) |
| 0.0 |
0.1 |
GO:0002541 |
activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 |
0.7 |
GO:0015701 |
bicarbonate transport(GO:0015701) |
| 0.0 |
0.2 |
GO:0007258 |
JUN phosphorylation(GO:0007258) |
| 0.0 |
0.1 |
GO:0071425 |
hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 |
0.2 |
GO:0019509 |
L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 |
0.4 |
GO:0001503 |
ossification(GO:0001503) |
| 0.0 |
0.1 |
GO:0007217 |
tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 |
0.3 |
GO:0060644 |
mammary gland epithelial cell differentiation(GO:0060644) |
| 0.0 |
0.9 |
GO:0042771 |
intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 |
1.7 |
GO:0006968 |
cellular defense response(GO:0006968) |
| 0.0 |
0.4 |
GO:0051966 |
regulation of synaptic transmission, glutamatergic(GO:0051966) |
| 0.0 |
0.0 |
GO:0048791 |
calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 |
0.6 |
GO:0050806 |
positive regulation of synaptic transmission(GO:0050806) |
| 0.0 |
0.2 |
GO:0016045 |
detection of bacterium(GO:0016045) |
| 0.0 |
0.4 |
GO:0046777 |
protein autophosphorylation(GO:0046777) |
| 0.0 |
0.2 |
GO:0016576 |
histone dephosphorylation(GO:0016576) |
| 0.0 |
0.4 |
GO:0071300 |
cellular response to retinoic acid(GO:0071300) |
| 0.0 |
2.2 |
GO:0007204 |
positive regulation of cytosolic calcium ion concentration(GO:0007204) |
| 0.0 |
0.5 |
GO:0045907 |
positive regulation of vasoconstriction(GO:0045907) |
| 0.0 |
4.5 |
GO:0006470 |
protein dephosphorylation(GO:0006470) |
| 0.0 |
0.2 |
GO:0016180 |
snRNA processing(GO:0016180) |
| 0.0 |
0.3 |
GO:0007168 |
receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 |
0.5 |
GO:0007528 |
neuromuscular junction development(GO:0007528) |
| 0.0 |
0.8 |
GO:0000080 |
mitotic G1 phase(GO:0000080) |
| 0.0 |
0.2 |
GO:0030262 |
apoptotic nuclear changes(GO:0030262) |
| 0.0 |
0.8 |
GO:0006730 |
one-carbon metabolic process(GO:0006730) |
| 0.0 |
0.7 |
GO:0007045 |
cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) |
| 0.0 |
1.0 |
GO:0070979 |
protein K11-linked ubiquitination(GO:0070979) |
| 0.0 |
0.1 |
GO:0043650 |
glutamate biosynthetic process(GO:0006537) glutamate catabolic process(GO:0006538) dicarboxylic acid biosynthetic process(GO:0043650) |
| 0.0 |
0.8 |
GO:0000186 |
activation of MAPKK activity(GO:0000186) |
| 0.0 |
0.4 |
GO:0007219 |
Notch signaling pathway(GO:0007219) |
| 0.0 |
0.0 |
GO:0048840 |
otolith development(GO:0048840) |
| 0.0 |
0.2 |
GO:0000188 |
inactivation of MAPK activity(GO:0000188) |
| 0.0 |
0.1 |
GO:0051001 |
negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.0 |
0.1 |
GO:0006642 |
triglyceride mobilization(GO:0006642) |
| 0.0 |
0.4 |
GO:0019098 |
mating behavior(GO:0007617) reproductive behavior(GO:0019098) multi-organism reproductive behavior(GO:0044705) |
| 0.0 |
1.7 |
GO:0007605 |
sensory perception of sound(GO:0007605) |
| 0.0 |
1.6 |
GO:0008277 |
regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 |
0.8 |
GO:0019233 |
sensory perception of pain(GO:0019233) |
| 0.0 |
0.1 |
GO:0046520 |
diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) sphingoid biosynthetic process(GO:0046520) |
| 0.0 |
0.2 |
GO:0001754 |
eye photoreceptor cell differentiation(GO:0001754) eye photoreceptor cell development(GO:0042462) |
| 0.0 |
0.2 |
GO:0021680 |
cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 |
0.6 |
GO:0016445 |
somatic diversification of immunoglobulins(GO:0016445) |
| 0.0 |
0.2 |
GO:0021549 |
cerebellum development(GO:0021549) |
| 0.0 |
0.2 |
GO:0033189 |
response to vitamin A(GO:0033189) |
| 0.0 |
0.0 |
GO:0045672 |
positive regulation of osteoclast differentiation(GO:0045672) |
| 0.0 |
0.5 |
GO:0006611 |
protein export from nucleus(GO:0006611) |
| 0.0 |
0.5 |
GO:0001892 |
embryonic placenta development(GO:0001892) |
| 0.0 |
0.6 |
GO:0000245 |
spliceosomal complex assembly(GO:0000245) |
| 0.0 |
0.2 |
GO:0098657 |
neurotransmitter uptake(GO:0001504) import into cell(GO:0098657) |
| 0.0 |
0.1 |
GO:0007635 |
chemosensory behavior(GO:0007635) |
| 0.0 |
1.0 |
GO:0006821 |
chloride transport(GO:0006821) |
| 0.0 |
0.2 |
GO:0008206 |
bile acid metabolic process(GO:0008206) |
| 0.0 |
1.8 |
GO:0045333 |
cellular respiration(GO:0045333) |
| 0.0 |
0.6 |
GO:0000289 |
nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 |
0.1 |
GO:0001960 |
negative regulation of cytokine-mediated signaling pathway(GO:0001960) |
| 0.0 |
0.2 |
GO:0007032 |
endosome organization(GO:0007032) |
| 0.0 |
0.8 |
GO:0061025 |
membrane fusion(GO:0061025) |
| 0.0 |
0.3 |
GO:0070830 |
bicellular tight junction assembly(GO:0070830) |
| 0.0 |
0.2 |
GO:0031293 |
membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 |
0.5 |
GO:1901342 |
regulation of angiogenesis(GO:0045765) regulation of vasculature development(GO:1901342) |
| 0.0 |
0.2 |
GO:0001501 |
skeletal system development(GO:0001501) |
| 0.0 |
0.2 |
GO:0015695 |
organic cation transport(GO:0015695) |
| 0.0 |
1.5 |
GO:0007565 |
female pregnancy(GO:0007565) |
| 0.0 |
0.5 |
GO:0042593 |
carbohydrate homeostasis(GO:0033500) glucose homeostasis(GO:0042593) |
| 0.0 |
1.3 |
GO:0008654 |
phospholipid biosynthetic process(GO:0008654) |
| 0.0 |
0.2 |
GO:0006596 |
polyamine biosynthetic process(GO:0006596) |
| 0.0 |
1.3 |
GO:0006367 |
transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 |
0.9 |
GO:0006338 |
chromatin remodeling(GO:0006338) |
| 0.0 |
0.6 |
GO:0050852 |
T cell receptor signaling pathway(GO:0050852) |
| 0.0 |
0.1 |
GO:0030382 |
sperm mitochondrion organization(GO:0030382) |
| 0.0 |
0.6 |
GO:0030216 |
keratinocyte differentiation(GO:0030216) |
| 0.0 |
0.2 |
GO:0000768 |
syncytium formation by plasma membrane fusion(GO:0000768) myoblast fusion(GO:0007520) |
| 0.0 |
0.2 |
GO:0051865 |
protein autoubiquitination(GO:0051865) |
| 0.0 |
0.7 |
GO:0016579 |
protein deubiquitination(GO:0016579) |
| 0.0 |
0.5 |
GO:0016064 |
immunoglobulin mediated immune response(GO:0016064) |
| 0.0 |
0.0 |
GO:0019886 |
antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 |
0.1 |
GO:0007212 |
dopamine receptor signaling pathway(GO:0007212) |
| 0.0 |
0.2 |
GO:0006000 |
fructose metabolic process(GO:0006000) |
| 0.0 |
1.5 |
GO:0007268 |
chemical synaptic transmission(GO:0007268) anterograde trans-synaptic signaling(GO:0098916) trans-synaptic signaling(GO:0099537) |
| 0.0 |
0.2 |
GO:0006098 |
pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.0 |
0.2 |
GO:0030514 |
negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 |
0.3 |
GO:0006469 |
negative regulation of protein kinase activity(GO:0006469) |