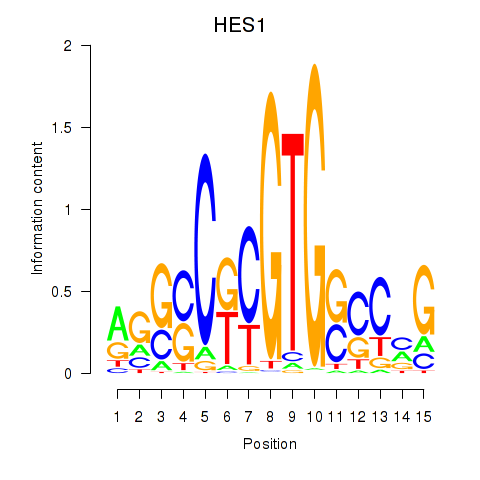
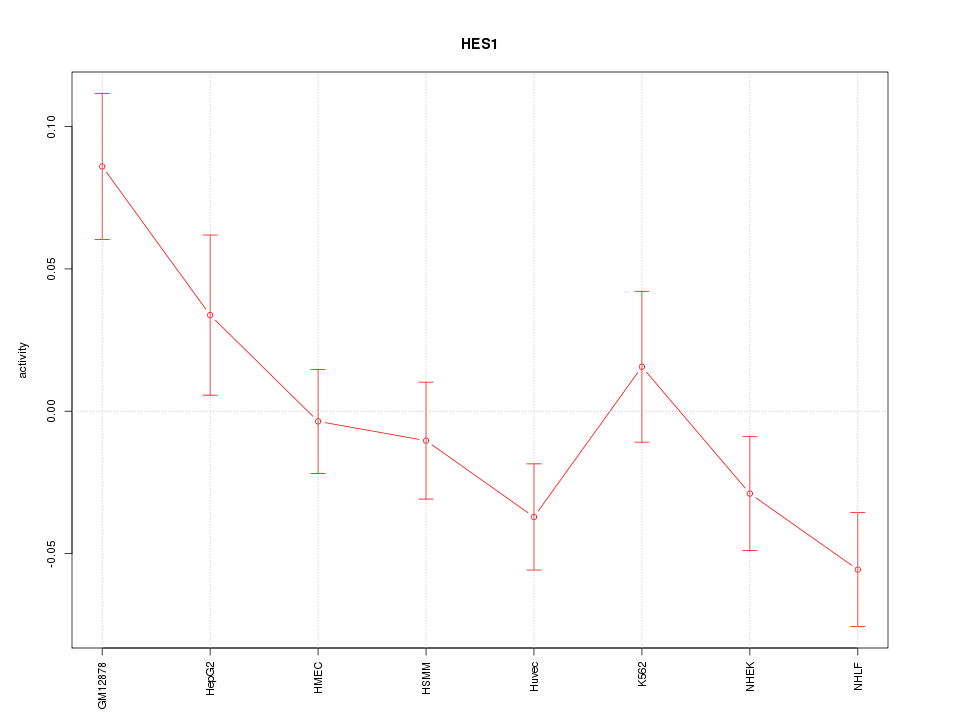
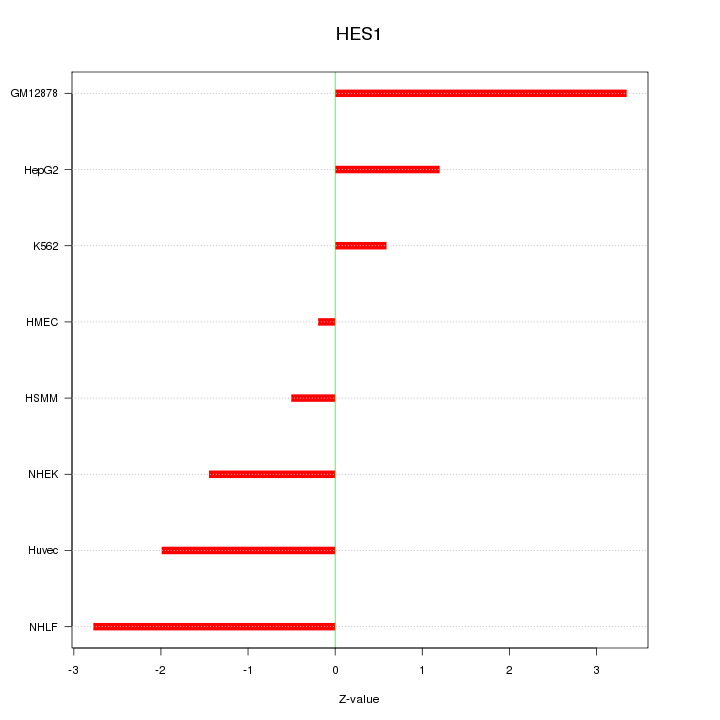
| 1.6 |
4.9 |
GO:0000738 |
DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.9 |
2.8 |
GO:0019242 |
methylglyoxal biosynthetic process(GO:0019242) |
| 0.6 |
1.8 |
GO:0015920 |
lipopolysaccharide transport(GO:0015920) |
| 0.4 |
1.3 |
GO:0046102 |
inosine metabolic process(GO:0046102) |
| 0.4 |
1.3 |
GO:0061117 |
negative regulation of cardiac muscle tissue growth(GO:0055022) negative regulation of cardiac muscle tissue development(GO:0055026) negative regulation of cardiac muscle cell proliferation(GO:0060044) negative regulation of heart growth(GO:0061117) |
| 0.4 |
1.2 |
GO:0043095 |
regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.3 |
1.0 |
GO:0071848 |
regulation of fever generation by regulation of prostaglandin secretion(GO:0071810) positive regulation of fever generation by positive regulation of prostaglandin secretion(GO:0071812) TNFSF11-mediated signaling pathway(GO:0071847) positive regulation of ERK1 and ERK2 cascade via TNFSF11-mediated signaling(GO:0071848) regulation of fever generation by prostaglandin secretion(GO:0100009) |
| 0.3 |
0.9 |
GO:0018916 |
nitrobenzene metabolic process(GO:0018916) |
| 0.3 |
0.9 |
GO:0046015 |
carbon catabolite regulation of transcription(GO:0045990) regulation of transcription by glucose(GO:0046015) positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.2 |
0.7 |
GO:0032927 |
positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.2 |
0.2 |
GO:0016458 |
gene silencing(GO:0016458) |
| 0.2 |
1.4 |
GO:0045654 |
positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.2 |
0.7 |
GO:0006273 |
lagging strand elongation(GO:0006273) |
| 0.2 |
0.9 |
GO:0033197 |
response to vitamin E(GO:0033197) |
| 0.2 |
0.7 |
GO:0006097 |
glyoxylate cycle(GO:0006097) |
| 0.2 |
0.7 |
GO:0019919 |
pathogenesis(GO:0009405) peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.2 |
0.6 |
GO:0061298 |
retina vasculature development in camera-type eye(GO:0061298) retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.2 |
0.6 |
GO:0048496 |
maintenance of organ identity(GO:0048496) |
| 0.2 |
0.8 |
GO:0009048 |
dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.2 |
1.4 |
GO:0045007 |
depurination(GO:0045007) |
| 0.2 |
0.6 |
GO:0021754 |
facial nucleus development(GO:0021754) cell proliferation in midbrain(GO:0033278) |
| 0.2 |
2.2 |
GO:0035590 |
purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.2 |
1.2 |
GO:0033152 |
immunoglobulin V(D)J recombination(GO:0033152) |
| 0.2 |
0.6 |
GO:0007206 |
phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.2 |
1.0 |
GO:0070935 |
3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.2 |
0.6 |
GO:0003011 |
diaphragm contraction(GO:0002086) involuntary skeletal muscle contraction(GO:0003011) |
| 0.2 |
0.9 |
GO:0003093 |
regulation of glomerular filtration(GO:0003093) glomerular filtration(GO:0003094) renal filtration(GO:0097205) regulation of renal system process(GO:0098801) |
| 0.2 |
3.4 |
GO:0006386 |
transcription elongation from RNA polymerase III promoter(GO:0006385) termination of RNA polymerase III transcription(GO:0006386) |
| 0.2 |
0.5 |
GO:0070544 |
histone H3-K36 demethylation(GO:0070544) |
| 0.2 |
1.2 |
GO:0070779 |
D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.2 |
0.5 |
GO:1901419 |
regulation of response to alcohol(GO:1901419) |
| 0.2 |
0.8 |
GO:0006857 |
oligopeptide transport(GO:0006857) |
| 0.2 |
0.6 |
GO:0034629 |
cellular protein complex localization(GO:0034629) |
| 0.2 |
0.5 |
GO:0002575 |
basophil chemotaxis(GO:0002575) |
| 0.2 |
0.3 |
GO:0031648 |
protein destabilization(GO:0031648) |
| 0.2 |
0.5 |
GO:0070601 |
maintenance of DNA methylation(GO:0010216) centromeric sister chromatid cohesion(GO:0070601) regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.1 |
1.3 |
GO:0007090 |
obsolete regulation of S phase of mitotic cell cycle(GO:0007090) |
| 0.1 |
0.4 |
GO:0045603 |
positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.1 |
0.8 |
GO:0071699 |
olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.1 |
0.4 |
GO:0043276 |
anoikis(GO:0043276) |
| 0.1 |
0.9 |
GO:0022027 |
interkinetic nuclear migration(GO:0022027) |
| 0.1 |
1.1 |
GO:0001706 |
endoderm formation(GO:0001706) |
| 0.1 |
0.5 |
GO:0002032 |
desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.1 |
0.2 |
GO:0061188 |
regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.1 |
1.1 |
GO:0048387 |
negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 |
0.4 |
GO:0035623 |
renal glucose absorption(GO:0035623) |
| 0.1 |
0.5 |
GO:0014040 |
regulation of Schwann cell differentiation(GO:0014038) positive regulation of Schwann cell differentiation(GO:0014040) |
| 0.1 |
0.6 |
GO:0010508 |
positive regulation of autophagy(GO:0010508) |
| 0.1 |
0.9 |
GO:0006188 |
IMP biosynthetic process(GO:0006188) IMP metabolic process(GO:0046040) |
| 0.1 |
0.4 |
GO:0000463 |
maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) maturation of LSU-rRNA(GO:0000470) |
| 0.1 |
0.9 |
GO:0070327 |
thyroid hormone transport(GO:0070327) |
| 0.1 |
0.6 |
GO:0006422 |
aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.1 |
0.5 |
GO:0044380 |
protein localization to cytoskeleton(GO:0044380) protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.1 |
0.3 |
GO:0006428 |
isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.1 |
0.4 |
GO:0060013 |
righting reflex(GO:0060013) |
| 0.1 |
0.3 |
GO:0000722 |
telomere maintenance via recombination(GO:0000722) |
| 0.1 |
0.5 |
GO:0045778 |
positive regulation of ossification(GO:0045778) |
| 0.1 |
0.2 |
GO:0060298 |
positive regulation of sarcomere organization(GO:0060298) |
| 0.1 |
1.0 |
GO:0007130 |
synaptonemal complex assembly(GO:0007130) |
| 0.1 |
0.9 |
GO:0032957 |
inositol trisphosphate metabolic process(GO:0032957) |
| 0.1 |
0.3 |
GO:0006288 |
base-excision repair, DNA ligation(GO:0006288) |
| 0.1 |
0.2 |
GO:0043654 |
recognition of apoptotic cell(GO:0043654) |
| 0.1 |
0.4 |
GO:0007182 |
common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.1 |
0.3 |
GO:0021819 |
layer formation in cerebral cortex(GO:0021819) |
| 0.1 |
0.4 |
GO:0017183 |
peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 |
0.3 |
GO:0060743 |
epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.1 |
0.2 |
GO:0021769 |
orbitofrontal cortex development(GO:0021769) ventricular zone neuroblast division(GO:0021847) fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) fibroblast growth factor receptor signaling pathway involved in orbitofrontal cortex development(GO:0035607) fibroblast growth factor receptor signaling pathway involved in mammary gland specification(GO:0060595) mammary gland bud formation(GO:0060615) branch elongation involved in salivary gland morphogenesis(GO:0060667) mesenchymal cell differentiation involved in lung development(GO:0060915) |
| 0.1 |
0.3 |
GO:0010940 |
positive regulation of necrotic cell death(GO:0010940) |
| 0.1 |
0.3 |
GO:0071894 |
histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.1 |
0.3 |
GO:0034219 |
carbohydrate transmembrane transport(GO:0034219) |
| 0.1 |
0.3 |
GO:0019985 |
translesion synthesis(GO:0019985) |
| 0.1 |
1.0 |
GO:0050858 |
negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 |
0.5 |
GO:0009820 |
alkaloid metabolic process(GO:0009820) |
| 0.1 |
0.2 |
GO:0014063 |
regulation of serotonin secretion(GO:0014062) negative regulation of serotonin secretion(GO:0014063) |
| 0.1 |
0.9 |
GO:0007076 |
mitotic chromosome condensation(GO:0007076) |
| 0.1 |
0.3 |
GO:0071294 |
cellular response to zinc ion(GO:0071294) |
| 0.1 |
0.3 |
GO:0042816 |
pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.1 |
0.3 |
GO:0019509 |
L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 |
0.3 |
GO:0010957 |
negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.1 |
0.3 |
GO:0032020 |
ISG15-protein conjugation(GO:0032020) |
| 0.1 |
0.2 |
GO:0001830 |
trophectodermal cell fate commitment(GO:0001830) |
| 0.1 |
2.3 |
GO:0006271 |
DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.1 |
0.2 |
GO:0000114 |
obsolete regulation of transcription involved in G1 phase of mitotic cell cycle(GO:0000114) |
| 0.1 |
1.0 |
GO:0051319 |
mitotic G2 phase(GO:0000085) G2 phase(GO:0051319) |
| 0.1 |
0.4 |
GO:0043457 |
regulation of cellular respiration(GO:0043457) |
| 0.1 |
0.2 |
GO:2000078 |
epithelial cilium movement(GO:0003351) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) regulation of type B pancreatic cell development(GO:2000074) positive regulation of type B pancreatic cell development(GO:2000078) |
| 0.1 |
0.3 |
GO:0001510 |
RNA methylation(GO:0001510) |
| 0.1 |
0.6 |
GO:0035235 |
ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 |
0.2 |
GO:0050804 |
modulation of synaptic transmission(GO:0050804) |
| 0.1 |
0.3 |
GO:0006686 |
sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 |
0.6 |
GO:0033261 |
obsolete regulation of S phase(GO:0033261) |
| 0.1 |
0.5 |
GO:0015886 |
heme transport(GO:0015886) |
| 0.1 |
0.3 |
GO:0035405 |
histone-threonine phosphorylation(GO:0035405) |
| 0.0 |
0.1 |
GO:0032481 |
positive regulation of type I interferon production(GO:0032481) positive regulation of interferon-alpha production(GO:0032727) |
| 0.0 |
0.2 |
GO:0006384 |
transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 |
4.5 |
GO:0006094 |
gluconeogenesis(GO:0006094) |
| 0.0 |
0.1 |
GO:0044314 |
protein K27-linked ubiquitination(GO:0044314) |
| 0.0 |
0.6 |
GO:0032411 |
positive regulation of transporter activity(GO:0032411) |
| 0.0 |
0.4 |
GO:0046831 |
regulation of nucleobase-containing compound transport(GO:0032239) regulation of RNA export from nucleus(GO:0046831) |
| 0.0 |
1.3 |
GO:0006783 |
heme biosynthetic process(GO:0006783) |
| 0.0 |
2.3 |
GO:0060041 |
retina development in camera-type eye(GO:0060041) |
| 0.0 |
0.3 |
GO:0051383 |
kinetochore assembly(GO:0051382) kinetochore organization(GO:0051383) |
| 0.0 |
0.3 |
GO:0000729 |
DNA double-strand break processing(GO:0000729) |
| 0.0 |
0.7 |
GO:0006400 |
tRNA modification(GO:0006400) |
| 0.0 |
0.3 |
GO:0033235 |
positive regulation of protein sumoylation(GO:0033235) |
| 0.0 |
0.7 |
GO:0050812 |
acetyl-CoA biosynthetic process from pyruvate(GO:0006086) regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 |
0.1 |
GO:0001941 |
postsynaptic membrane organization(GO:0001941) |
| 0.0 |
0.0 |
GO:1902116 |
negative regulation of organelle assembly(GO:1902116) |
| 0.0 |
0.8 |
GO:0032012 |
regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 |
0.9 |
GO:0030968 |
endoplasmic reticulum unfolded protein response(GO:0030968) cellular response to unfolded protein(GO:0034620) |
| 0.0 |
0.1 |
GO:0009134 |
nucleoside diphosphate catabolic process(GO:0009134) ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.0 |
0.1 |
GO:0021937 |
cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 0.0 |
0.1 |
GO:0051597 |
response to methylmercury(GO:0051597) |
| 0.0 |
0.2 |
GO:0006983 |
ER overload response(GO:0006983) |
| 0.0 |
0.7 |
GO:0032608 |
interferon-beta production(GO:0032608) regulation of interferon-beta production(GO:0032648) |
| 0.0 |
0.1 |
GO:0060318 |
CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) definitive erythrocyte differentiation(GO:0060318) |
| 0.0 |
0.1 |
GO:0070091 |
glucagon secretion(GO:0070091) regulation of glucagon secretion(GO:0070092) |
| 0.0 |
0.3 |
GO:0007143 |
female meiotic division(GO:0007143) |
| 0.0 |
1.4 |
GO:0042771 |
intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 |
0.6 |
GO:0007020 |
microtubule nucleation(GO:0007020) |
| 0.0 |
0.4 |
GO:0008054 |
negative regulation of cyclin-dependent protein serine/threonine kinase by cyclin degradation(GO:0008054) |
| 0.0 |
0.1 |
GO:2000051 |
negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 |
0.3 |
GO:1901019 |
regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) regulation of calcium ion transmembrane transporter activity(GO:1901019) regulation of cation channel activity(GO:2001257) |
| 0.0 |
0.3 |
GO:0008535 |
respiratory chain complex IV assembly(GO:0008535) |
| 0.0 |
0.1 |
GO:0071168 |
negative regulation of sister chromatid cohesion(GO:0045875) protein localization to chromatin(GO:0071168) |
| 0.0 |
0.1 |
GO:0015910 |
peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 |
0.2 |
GO:0006999 |
nuclear pore organization(GO:0006999) |
| 0.0 |
0.4 |
GO:0046037 |
phototransduction, visible light(GO:0007603) GMP metabolic process(GO:0046037) |
| 0.0 |
0.1 |
GO:0021684 |
cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 |
0.4 |
GO:0016226 |
iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 |
0.2 |
GO:0018027 |
peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 |
0.2 |
GO:0033137 |
negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 |
0.7 |
GO:0014823 |
response to activity(GO:0014823) |
| 0.0 |
0.2 |
GO:0034969 |
histone arginine methylation(GO:0034969) |
| 0.0 |
0.2 |
GO:0043545 |
Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 |
0.2 |
GO:0035082 |
axoneme assembly(GO:0035082) |
| 0.0 |
0.1 |
GO:0015919 |
peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 |
0.3 |
GO:1901984 |
negative regulation of histone acetylation(GO:0035067) negative regulation of protein acetylation(GO:1901984) negative regulation of peptidyl-lysine acetylation(GO:2000757) |
| 0.0 |
0.2 |
GO:0045075 |
interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 |
2.5 |
GO:0043010 |
camera-type eye development(GO:0043010) |
| 0.0 |
0.2 |
GO:0031427 |
response to methotrexate(GO:0031427) |
| 0.0 |
2.1 |
GO:0050680 |
negative regulation of epithelial cell proliferation(GO:0050680) |
| 0.0 |
0.4 |
GO:0043097 |
pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.0 |
0.1 |
GO:0000394 |
RNA splicing, via endonucleolytic cleavage and ligation(GO:0000394) tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 |
0.0 |
GO:0060732 |
positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 |
0.0 |
GO:0060992 |
response to fungicide(GO:0060992) |
| 0.0 |
0.3 |
GO:0007029 |
endoplasmic reticulum organization(GO:0007029) |
| 0.0 |
0.1 |
GO:0034720 |
histone H3-K4 demethylation(GO:0034720) |
| 0.0 |
0.7 |
GO:0000188 |
inactivation of MAPK activity(GO:0000188) |
| 0.0 |
0.1 |
GO:0048172 |
regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 |
0.3 |
GO:0046902 |
regulation of mitochondrial membrane permeability(GO:0046902) regulation of membrane permeability(GO:0090559) |
| 0.0 |
0.2 |
GO:0070934 |
CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 |
0.2 |
GO:0040032 |
post-embryonic body morphogenesis(GO:0040032) |
| 0.0 |
0.1 |
GO:0006896 |
Golgi to endosome transport(GO:0006895) Golgi to vacuole transport(GO:0006896) |
| 0.0 |
0.3 |
GO:0051051 |
negative regulation of transport(GO:0051051) |
| 0.0 |
3.1 |
GO:0006333 |
chromatin assembly or disassembly(GO:0006333) |
| 0.0 |
0.5 |
GO:0072331 |
signal transduction by p53 class mediator(GO:0072331) |
| 0.0 |
0.9 |
GO:0046777 |
protein autophosphorylation(GO:0046777) |
| 0.0 |
0.2 |
GO:0019885 |
antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 |
0.1 |
GO:0003416 |
endochondral bone growth(GO:0003416) growth plate cartilage development(GO:0003417) bone growth(GO:0098868) |
| 0.0 |
1.1 |
GO:0006369 |
termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 |
0.6 |
GO:0031214 |
biomineral tissue development(GO:0031214) |
| 0.0 |
0.2 |
GO:0046051 |
GTP biosynthetic process(GO:0006183) UTP biosynthetic process(GO:0006228) CTP biosynthetic process(GO:0006241) pyrimidine ribonucleoside triphosphate metabolic process(GO:0009208) pyrimidine ribonucleoside triphosphate biosynthetic process(GO:0009209) CTP metabolic process(GO:0046036) GTP metabolic process(GO:0046039) UTP metabolic process(GO:0046051) guanosine-containing compound biosynthetic process(GO:1901070) |
| 0.0 |
0.2 |
GO:0075713 |
establishment of viral latency(GO:0019043) establishment of integrated proviral latency(GO:0075713) |
| 0.0 |
0.1 |
GO:0045767 |
obsolete regulation of anti-apoptosis(GO:0045767) |
| 0.0 |
0.1 |
GO:0007023 |
post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 |
0.7 |
GO:0043966 |
histone H3 acetylation(GO:0043966) |
| 0.0 |
0.2 |
GO:0019221 |
cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 |
0.3 |
GO:0016254 |
preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 |
0.2 |
GO:0009086 |
methionine biosynthetic process(GO:0009086) |
| 0.0 |
0.1 |
GO:0006658 |
phosphatidylserine metabolic process(GO:0006658) |
| 0.0 |
0.4 |
GO:0007214 |
gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 |
0.3 |
GO:0019370 |
leukotriene biosynthetic process(GO:0019370) |
| 0.0 |
0.2 |
GO:0051928 |
positive regulation of calcium ion transport(GO:0051928) |
| 0.0 |
0.3 |
GO:0046834 |
lipid phosphorylation(GO:0046834) |
| 0.0 |
0.4 |
GO:0021675 |
nerve development(GO:0021675) |
| 0.0 |
0.3 |
GO:0098657 |
neurotransmitter uptake(GO:0001504) import into cell(GO:0098657) |
| 0.0 |
0.1 |
GO:0006600 |
creatine metabolic process(GO:0006600) |
| 0.0 |
0.3 |
GO:0042147 |
retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 |
0.7 |
GO:0048706 |
embryonic skeletal system development(GO:0048706) |
| 0.0 |
0.1 |
GO:0006378 |
mRNA polyadenylation(GO:0006378) |
| 0.0 |
0.0 |
GO:0045814 |
negative regulation of gene expression, epigenetic(GO:0045814) |
| 0.0 |
0.3 |
GO:0006353 |
DNA-templated transcription, termination(GO:0006353) |
| 0.0 |
0.2 |
GO:0006906 |
vesicle fusion(GO:0006906) |
| 0.0 |
0.6 |
GO:0000079 |
regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 |
0.1 |
GO:0045604 |
regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 |
0.1 |
GO:0061756 |
leukocyte tethering or rolling(GO:0050901) leukocyte adhesion to vascular endothelial cell(GO:0061756) |