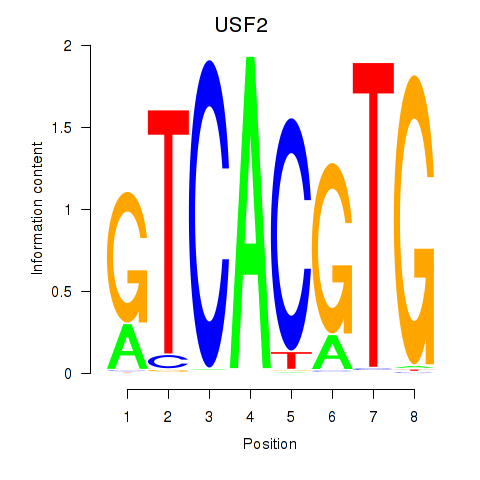
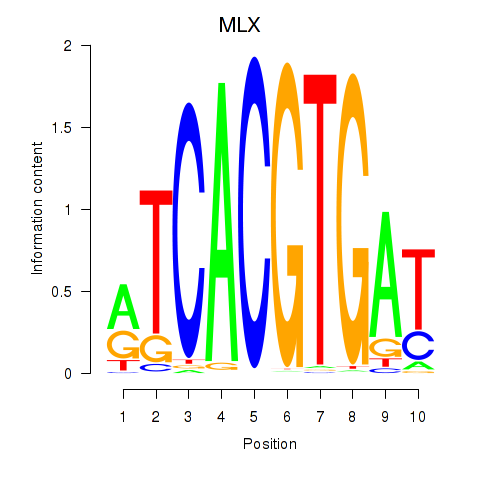
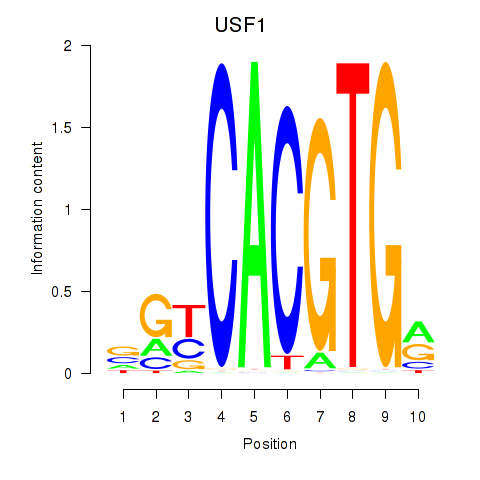
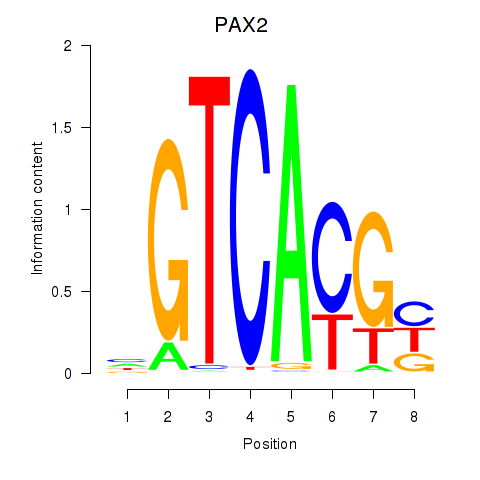
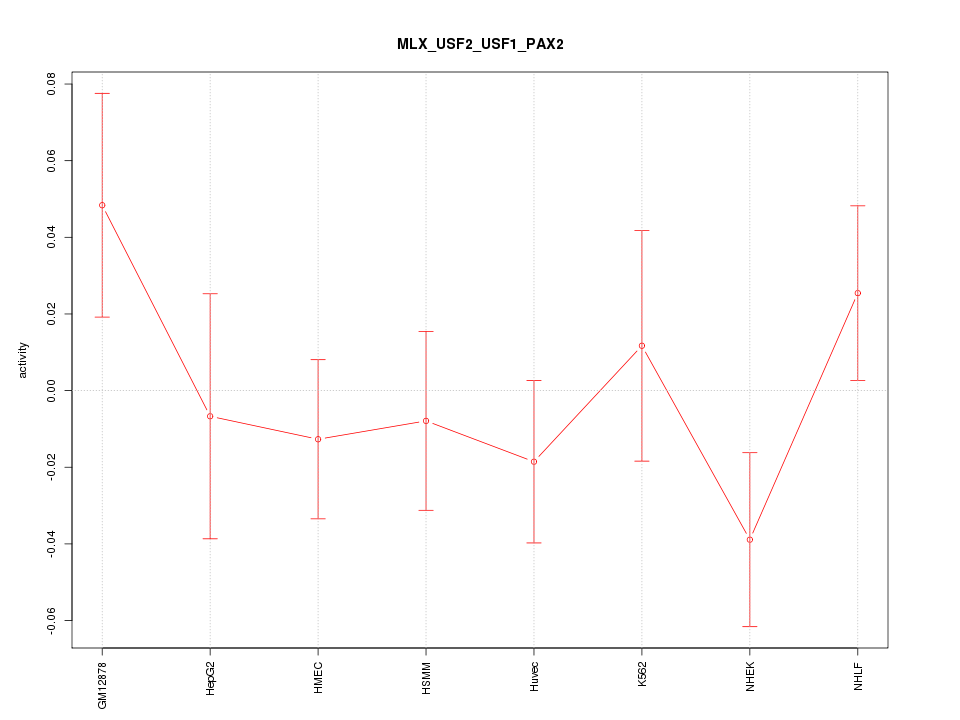
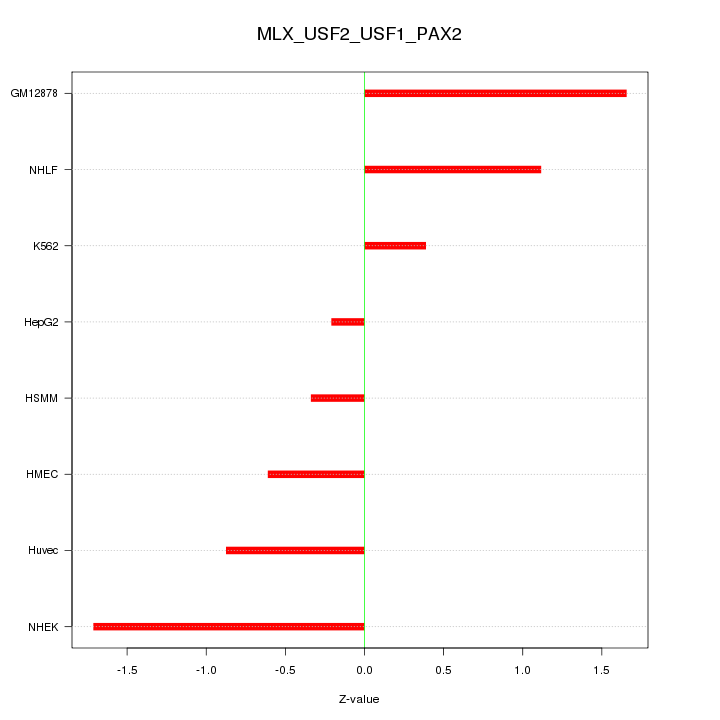
| 0.7 |
5.0 |
GO:0060155 |
platelet dense granule organization(GO:0060155) |
| 0.5 |
1.4 |
GO:0043371 |
negative regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043371) negative regulation of T-helper cell differentiation(GO:0045623) negative regulation of CD4-positive, alpha-beta T cell activation(GO:2000515) |
| 0.4 |
1.3 |
GO:0014022 |
neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.4 |
0.4 |
GO:0032497 |
detection of lipopolysaccharide(GO:0032497) |
| 0.3 |
1.0 |
GO:0070407 |
oxidation-dependent protein catabolic process(GO:0070407) |
| 0.3 |
0.9 |
GO:0000738 |
DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.3 |
1.5 |
GO:0010508 |
positive regulation of autophagy(GO:0010508) |
| 0.3 |
0.8 |
GO:0032071 |
regulation of endodeoxyribonuclease activity(GO:0032071) negative regulation of helicase activity(GO:0051097) |
| 0.3 |
0.8 |
GO:0010273 |
regulation of oxidative phosphorylation(GO:0002082) detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.2 |
1.0 |
GO:0032929 |
negative regulation of superoxide anion generation(GO:0032929) |
| 0.2 |
0.2 |
GO:0046449 |
allantoin metabolic process(GO:0000255) creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.2 |
1.5 |
GO:0002726 |
positive regulation of T cell cytokine production(GO:0002726) |
| 0.2 |
1.2 |
GO:0010832 |
negative regulation of myotube differentiation(GO:0010832) |
| 0.2 |
1.6 |
GO:0006689 |
ganglioside catabolic process(GO:0006689) |
| 0.2 |
0.6 |
GO:0007034 |
vacuolar transport(GO:0007034) |
| 0.2 |
0.4 |
GO:0009952 |
anterior/posterior pattern specification(GO:0009952) |
| 0.2 |
0.5 |
GO:0034139 |
regulation of toll-like receptor 2 signaling pathway(GO:0034135) regulation of toll-like receptor 3 signaling pathway(GO:0034139) negative regulation of epithelial cell apoptotic process(GO:1904036) negative regulation of endothelial cell apoptotic process(GO:2000352) |
| 0.2 |
0.7 |
GO:0044380 |
protein localization to cytoskeleton(GO:0044380) protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.2 |
0.9 |
GO:0035405 |
histone-threonine phosphorylation(GO:0035405) |
| 0.2 |
0.6 |
GO:0010157 |
response to chlorate(GO:0010157) |
| 0.2 |
0.9 |
GO:0043316 |
natural killer cell activation involved in immune response(GO:0002323) cytotoxic T cell degranulation(GO:0043316) natural killer cell degranulation(GO:0043320) |
| 0.2 |
1.2 |
GO:0033160 |
positive regulation of protein import into nucleus, translocation(GO:0033160) |
| 0.1 |
1.2 |
GO:0006188 |
IMP biosynthetic process(GO:0006188) IMP metabolic process(GO:0046040) |
| 0.1 |
1.4 |
GO:0015074 |
DNA integration(GO:0015074) |
| 0.1 |
2.0 |
GO:0070193 |
synaptonemal complex organization(GO:0070193) |
| 0.1 |
0.4 |
GO:0097237 |
cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.1 |
0.5 |
GO:0002689 |
negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.1 |
0.4 |
GO:0002887 |
negative regulation of myeloid leukocyte mediated immunity(GO:0002887) heme oxidation(GO:0006788) smooth muscle hyperplasia(GO:0014806) muscle hyperplasia(GO:0014900) negative regulation of leukocyte degranulation(GO:0043301) |
| 0.1 |
1.0 |
GO:0071354 |
cellular response to interleukin-6(GO:0071354) |
| 0.1 |
0.6 |
GO:0060259 |
regulation of feeding behavior(GO:0060259) |
| 0.1 |
0.5 |
GO:0035470 |
positive regulation of vascular wound healing(GO:0035470) |
| 0.1 |
0.3 |
GO:0010586 |
miRNA metabolic process(GO:0010586) miRNA catabolic process(GO:0010587) |
| 0.1 |
1.7 |
GO:0071378 |
growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 |
2.0 |
GO:0046655 |
folic acid metabolic process(GO:0046655) |
| 0.1 |
0.4 |
GO:0035283 |
central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 |
0.3 |
GO:0051066 |
dihydrobiopterin metabolic process(GO:0051066) |
| 0.1 |
0.3 |
GO:0070734 |
histone H3-K27 methylation(GO:0070734) |
| 0.1 |
1.6 |
GO:0071230 |
cellular response to amino acid stimulus(GO:0071230) |
| 0.1 |
0.4 |
GO:0043653 |
mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 |
0.5 |
GO:0032959 |
inositol trisphosphate biosynthetic process(GO:0032959) positive regulation of calcium ion transmembrane transporter activity(GO:1901021) positive regulation of cation channel activity(GO:2001259) |
| 0.1 |
0.6 |
GO:0045176 |
apical protein localization(GO:0045176) |
| 0.1 |
0.2 |
GO:0070171 |
negative regulation of tooth mineralization(GO:0070171) |
| 0.1 |
0.2 |
GO:0034626 |
fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 |
0.8 |
GO:0032355 |
response to estradiol(GO:0032355) |
| 0.1 |
0.2 |
GO:0006419 |
alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 |
0.9 |
GO:0006268 |
DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 |
0.2 |
GO:0034755 |
iron ion transmembrane transport(GO:0034755) |
| 0.1 |
1.1 |
GO:0007253 |
cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.1 |
0.5 |
GO:0045008 |
depyrimidination(GO:0045008) |
| 0.1 |
1.5 |
GO:0031055 |
DNA replication-independent nucleosome assembly(GO:0006336) chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) DNA replication-independent nucleosome organization(GO:0034724) CENP-A containing chromatin organization(GO:0061641) |
| 0.1 |
0.7 |
GO:0046037 |
GMP metabolic process(GO:0046037) |
| 0.1 |
1.9 |
GO:0007157 |
heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 |
0.1 |
GO:0071504 |
response to heparin(GO:0071503) cellular response to heparin(GO:0071504) |
| 0.1 |
0.3 |
GO:0042483 |
negative regulation of odontogenesis(GO:0042483) |
| 0.1 |
0.2 |
GO:0070384 |
Harderian gland development(GO:0070384) |
| 0.1 |
0.2 |
GO:0019858 |
cytosine metabolic process(GO:0019858) |
| 0.1 |
0.6 |
GO:0034638 |
phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 |
1.4 |
GO:0071577 |
zinc II ion transmembrane transport(GO:0071577) |
| 0.1 |
0.4 |
GO:0043985 |
histone H4-R3 methylation(GO:0043985) |
| 0.1 |
0.7 |
GO:0032703 |
negative regulation of interleukin-2 production(GO:0032703) |
| 0.1 |
0.2 |
GO:0071312 |
cellular response to alkaloid(GO:0071312) cellular response to caffeine(GO:0071313) |
| 0.1 |
0.5 |
GO:0060009 |
Sertoli cell development(GO:0060009) |
| 0.1 |
0.5 |
GO:0006069 |
ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.1 |
0.2 |
GO:0001575 |
globoside metabolic process(GO:0001575) |
| 0.1 |
0.2 |
GO:0048935 |
peripheral nervous system neuron development(GO:0048935) |
| 0.1 |
0.8 |
GO:0006260 |
DNA replication(GO:0006260) |
| 0.1 |
0.2 |
GO:0033262 |
regulation of nuclear cell cycle DNA replication(GO:0033262) |
| 0.1 |
0.3 |
GO:0009649 |
entrainment of circadian clock(GO:0009649) |
| 0.1 |
1.1 |
GO:0008283 |
cell proliferation(GO:0008283) |
| 0.0 |
1.0 |
GO:0016254 |
preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 |
0.1 |
GO:0090073 |
positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 |
1.4 |
GO:0006099 |
tricarboxylic acid cycle(GO:0006099) |
| 0.0 |
0.1 |
GO:0034498 |
early endosome to Golgi transport(GO:0034498) |
| 0.0 |
0.1 |
GO:0017014 |
protein nitrosylation(GO:0017014) peptidyl-cysteine S-nitrosylation(GO:0018119) |
| 0.0 |
0.1 |
GO:0021898 |
commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 |
2.2 |
GO:0048146 |
positive regulation of fibroblast proliferation(GO:0048146) |
| 0.0 |
0.1 |
GO:0014063 |
regulation of serotonin secretion(GO:0014062) negative regulation of serotonin secretion(GO:0014063) |
| 0.0 |
0.6 |
GO:0046827 |
positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 |
0.5 |
GO:0032876 |
regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 |
0.1 |
GO:0043441 |
acetoacetic acid biosynthetic process(GO:0043441) |
| 0.0 |
0.2 |
GO:0032933 |
SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 |
0.1 |
GO:0042339 |
keratan sulfate metabolic process(GO:0042339) |
| 0.0 |
0.1 |
GO:0006382 |
adenosine to inosine editing(GO:0006382) |
| 0.0 |
0.3 |
GO:0006975 |
DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 |
0.3 |
GO:0045039 |
protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 |
0.2 |
GO:0007182 |
common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.0 |
0.1 |
GO:0010216 |
maintenance of DNA methylation(GO:0010216) |
| 0.0 |
0.1 |
GO:0001833 |
inner cell mass cell proliferation(GO:0001833) |
| 0.0 |
0.5 |
GO:0006044 |
N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 |
0.9 |
GO:0030225 |
macrophage differentiation(GO:0030225) |
| 0.0 |
0.1 |
GO:0006655 |
phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin metabolic process(GO:0032048) cardiolipin biosynthetic process(GO:0032049) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.0 |
0.4 |
GO:0034643 |
establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 |
0.9 |
GO:0051865 |
protein autoubiquitination(GO:0051865) |
| 0.0 |
0.2 |
GO:0070245 |
positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.0 |
0.4 |
GO:0045070 |
positive regulation of viral genome replication(GO:0045070) |
| 0.0 |
0.1 |
GO:0048193 |
Golgi vesicle transport(GO:0048193) |
| 0.0 |
0.1 |
GO:0009113 |
purine nucleobase biosynthetic process(GO:0009113) |
| 0.0 |
0.2 |
GO:0019227 |
neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 |
0.1 |
GO:0015917 |
aminophospholipid transport(GO:0015917) |
| 0.0 |
0.1 |
GO:0009162 |
deoxyribonucleoside monophosphate metabolic process(GO:0009162) pyrimidine deoxyribonucleoside monophosphate metabolic process(GO:0009176) transformation of host cell by virus(GO:0019087) dUMP metabolic process(GO:0046078) |
| 0.0 |
0.1 |
GO:0060736 |
prostate gland growth(GO:0060736) |
| 0.0 |
0.1 |
GO:0042756 |
drinking behavior(GO:0042756) |
| 0.0 |
0.1 |
GO:0015670 |
carbon dioxide transport(GO:0015670) |
| 0.0 |
0.1 |
GO:0019482 |
beta-alanine metabolic process(GO:0019482) |
| 0.0 |
0.2 |
GO:0031664 |
regulation of lipopolysaccharide-mediated signaling pathway(GO:0031664) negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 |
1.1 |
GO:0016925 |
protein sumoylation(GO:0016925) |
| 0.0 |
0.2 |
GO:0032776 |
DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 |
0.3 |
GO:0060665 |
regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 |
0.1 |
GO:0044314 |
protein K27-linked ubiquitination(GO:0044314) |
| 0.0 |
0.3 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 |
0.2 |
GO:0034616 |
response to laminar fluid shear stress(GO:0034616) |
| 0.0 |
0.1 |
GO:0002092 |
positive regulation of receptor internalization(GO:0002092) |
| 0.0 |
0.3 |
GO:0045898 |
regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 |
0.3 |
GO:0015937 |
coenzyme A biosynthetic process(GO:0015937) |
| 0.0 |
0.2 |
GO:0032836 |
glomerular basement membrane development(GO:0032836) |
| 0.0 |
0.2 |
GO:0006072 |
glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 |
0.1 |
GO:0051534 |
negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 |
0.2 |
GO:0007217 |
tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 |
0.3 |
GO:0043084 |
penile erection(GO:0043084) |
| 0.0 |
1.0 |
GO:0006270 |
DNA replication initiation(GO:0006270) |
| 0.0 |
0.1 |
GO:0071344 |
diphosphate metabolic process(GO:0071344) |
| 0.0 |
0.3 |
GO:0045351 |
type I interferon biosynthetic process(GO:0045351) |
| 0.0 |
0.8 |
GO:0050830 |
defense response to Gram-positive bacterium(GO:0050830) |
| 0.0 |
0.2 |
GO:0008608 |
attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.0 |
0.3 |
GO:0031648 |
protein destabilization(GO:0031648) |
| 0.0 |
0.3 |
GO:0070389 |
chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 |
0.0 |
GO:0002268 |
follicular dendritic cell activation(GO:0002266) follicular dendritic cell differentiation(GO:0002268) |
| 0.0 |
0.0 |
GO:0034244 |
negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 |
1.9 |
GO:0006096 |
glycolytic process(GO:0006096) ATP generation from ADP(GO:0006757) |
| 0.0 |
1.2 |
GO:0030183 |
B cell differentiation(GO:0030183) |
| 0.0 |
0.1 |
GO:0022417 |
protein maturation by protein folding(GO:0022417) |
| 0.0 |
0.3 |
GO:0033169 |
histone H3-K9 demethylation(GO:0033169) |
| 0.0 |
0.3 |
GO:0006400 |
tRNA modification(GO:0006400) |
| 0.0 |
0.1 |
GO:0045007 |
base-excision repair, AP site formation(GO:0006285) depurination(GO:0045007) |
| 0.0 |
0.2 |
GO:0016601 |
Rac protein signal transduction(GO:0016601) |
| 0.0 |
0.0 |
GO:0007538 |
primary sex determination(GO:0007538) |
| 0.0 |
0.2 |
GO:0046831 |
regulation of RNA export from nucleus(GO:0046831) |
| 0.0 |
0.1 |
GO:0006726 |
eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) eye pigmentation(GO:0048069) |
| 0.0 |
0.2 |
GO:0023041 |
neuronal signal transduction(GO:0023041) |
| 0.0 |
0.1 |
GO:0071168 |
protein localization to chromatin(GO:0071168) |
| 0.0 |
0.2 |
GO:0031115 |
negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 |
0.9 |
GO:0019915 |
lipid storage(GO:0019915) |
| 0.0 |
0.0 |
GO:0032914 |
positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 |
0.4 |
GO:0006884 |
cell volume homeostasis(GO:0006884) |
| 0.0 |
0.0 |
GO:0007621 |
negative regulation of female receptivity(GO:0007621) female courtship behavior(GO:0008050) |
| 0.0 |
0.1 |
GO:0008089 |
anterograde axonal transport(GO:0008089) |
| 0.0 |
0.2 |
GO:0043523 |
regulation of neuron apoptotic process(GO:0043523) regulation of neuron death(GO:1901214) |
| 0.0 |
0.1 |
GO:0045618 |
positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.0 |
0.1 |
GO:0000076 |
DNA replication checkpoint(GO:0000076) |
| 0.0 |
0.4 |
GO:0033138 |
positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 |
2.0 |
GO:0008277 |
regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 |
0.1 |
GO:0032897 |
negative regulation of viral transcription(GO:0032897) |
| 0.0 |
0.1 |
GO:0060748 |
tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 |
0.4 |
GO:0000060 |
protein import into nucleus, translocation(GO:0000060) |
| 0.0 |
0.2 |
GO:0045446 |
endothelium development(GO:0003158) endothelial cell differentiation(GO:0045446) |
| 0.0 |
0.2 |
GO:0048070 |
regulation of developmental pigmentation(GO:0048070) |
| 0.0 |
0.3 |
GO:0030260 |
entry into host cell(GO:0030260) entry into host(GO:0044409) entry into cell of other organism involved in symbiotic interaction(GO:0051806) entry into other organism involved in symbiotic interaction(GO:0051828) |
| 0.0 |
0.4 |
GO:0006656 |
phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 |
1.0 |
GO:0006369 |
termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 |
0.3 |
GO:0006613 |
cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 |
0.1 |
GO:0046902 |
regulation of mitochondrial membrane permeability(GO:0046902) regulation of membrane permeability(GO:0090559) |
| 0.0 |
0.1 |
GO:0046112 |
nucleobase biosynthetic process(GO:0046112) |
| 0.0 |
0.1 |
GO:0002566 |
somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 |
0.0 |
GO:0032695 |
negative regulation of interleukin-12 production(GO:0032695) |
| 0.0 |
0.0 |
GO:0043438 |
acetoacetic acid metabolic process(GO:0043438) |
| 0.0 |
0.1 |
GO:0042816 |
pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 |
0.0 |
GO:0090047 |
obsolete positive regulation of transcription regulator activity(GO:0090047) |
| 0.0 |
0.0 |
GO:0043247 |
telomere capping(GO:0016233) protection from non-homologous end joining at telomere(GO:0031848) telomere maintenance in response to DNA damage(GO:0043247) |
| 0.0 |
0.1 |
GO:0032508 |
DNA duplex unwinding(GO:0032508) |
| 0.0 |
0.2 |
GO:0006098 |
pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.0 |
0.1 |
GO:0070934 |
CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 |
0.2 |
GO:0046839 |
phospholipid dephosphorylation(GO:0046839) |
| 0.0 |
0.2 |
GO:0006359 |
regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 |
0.1 |
GO:0018106 |
peptidyl-histidine phosphorylation(GO:0018106) |
| 0.0 |
0.0 |
GO:0048865 |
stem cell fate commitment(GO:0048865) stem cell fate determination(GO:0048867) |
| 0.0 |
0.1 |
GO:0006561 |
proline biosynthetic process(GO:0006561) |
| 0.0 |
0.1 |
GO:0033151 |
V(D)J recombination(GO:0033151) |
| 0.0 |
0.1 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 |
0.1 |
GO:0051013 |
microtubule severing(GO:0051013) |
| 0.0 |
0.4 |
GO:0008284 |
positive regulation of cell proliferation(GO:0008284) |
| 0.0 |
1.7 |
GO:0006333 |
chromatin assembly or disassembly(GO:0006333) |
| 0.0 |
0.1 |
GO:0006552 |
leucine catabolic process(GO:0006552) coenzyme A metabolic process(GO:0015936) |
| 0.0 |
0.4 |
GO:0051225 |
spindle assembly(GO:0051225) |
| 0.0 |
0.1 |
GO:0007021 |
tubulin complex assembly(GO:0007021) acetylcholine transport(GO:0015870) acetate ester transport(GO:1901374) |
| 0.0 |
0.1 |
GO:0008340 |
determination of adult lifespan(GO:0008340) |
| 0.0 |
0.2 |
GO:0046854 |
phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 |
0.3 |
GO:0045739 |
positive regulation of DNA repair(GO:0045739) |
| 0.0 |
0.1 |
GO:0090005 |
negative regulation of establishment of protein localization to plasma membrane(GO:0090005) negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.0 |
1.0 |
GO:0006521 |
regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.0 |
0.1 |
GO:0052646 |
alditol phosphate metabolic process(GO:0052646) |
| 0.0 |
0.8 |
GO:0006892 |
post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 |
0.2 |
GO:0007520 |
myoblast fusion(GO:0007520) |
| 0.0 |
0.3 |
GO:0034199 |
activation of protein kinase A activity(GO:0034199) |
| 0.0 |
4.3 |
GO:0051056 |
regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 |
0.2 |
GO:0006386 |
transcription elongation from RNA polymerase III promoter(GO:0006385) termination of RNA polymerase III transcription(GO:0006386) |
| 0.0 |
0.0 |
GO:0031133 |
regulation of axon diameter(GO:0031133) regulation of cell projection size(GO:0032536) |
| 0.0 |
0.0 |
GO:0003093 |
regulation of glomerular filtration(GO:0003093) glomerular filtration(GO:0003094) positive regulation of nucleobase-containing compound transport(GO:0032241) negative regulation of vasodilation(GO:0045908) regulation of mucus secretion(GO:0070255) negative regulation of mucus secretion(GO:0070256) renal filtration(GO:0097205) regulation of renal system process(GO:0098801) |
| 0.0 |
0.1 |
GO:0050679 |
positive regulation of epithelial cell proliferation(GO:0050679) |
| 0.0 |
0.2 |
GO:0000083 |
regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 |
0.0 |
GO:0000463 |
maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) maturation of LSU-rRNA(GO:0000470) |
| 0.0 |
0.5 |
GO:0006888 |
ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 |
0.1 |
GO:0019509 |
L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 |
0.1 |
GO:0007204 |
positive regulation of cytosolic calcium ion concentration(GO:0007204) |
| 0.0 |
0.2 |
GO:0043496 |
regulation of protein homodimerization activity(GO:0043496) |
| 0.0 |
0.2 |
GO:0014823 |
response to activity(GO:0014823) |
| 0.0 |
0.1 |
GO:0048251 |
elastic fiber assembly(GO:0048251) extracellular matrix assembly(GO:0085029) |