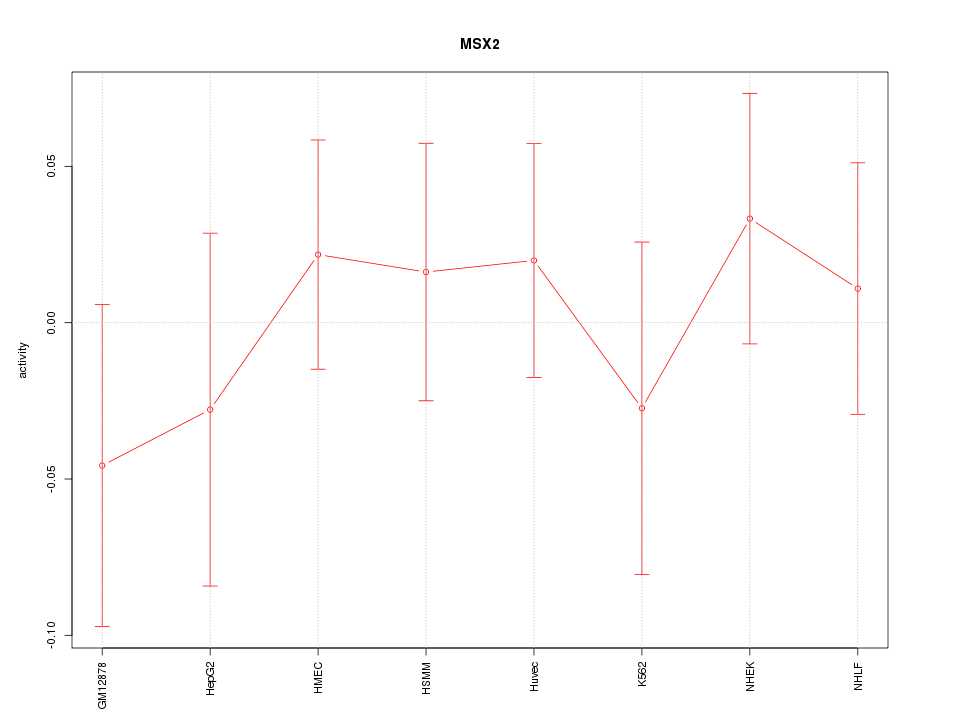
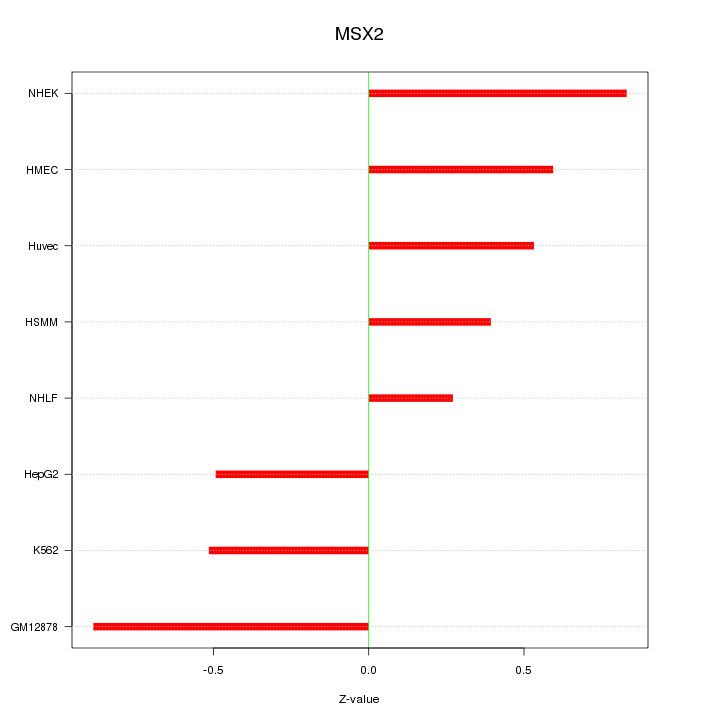
Motif ID: MSX2
Z-value: 0.597
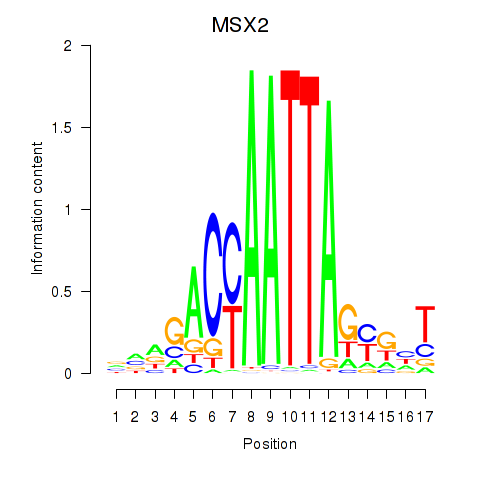
Transcription factors associated with MSX2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| MSX2 | ENSG00000120149.7 | MSX2 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.3 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.8 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.1 | GO:1904358 | positive regulation of telomere maintenance via telomerase(GO:0032212) positive regulation of telomere maintenance via telomere lengthening(GO:1904358) |
| 0.0 | 1.8 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 0.1 | GO:1902414 | protein localization to adherens junction(GO:0071896) protein localization to cell junction(GO:1902414) |
| 0.0 | 0.1 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 0.4 | GO:0048821 | erythrocyte development(GO:0048821) myeloid cell development(GO:0061515) |
| 0.0 | 0.3 | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0070059) |
| 0.0 | 0.2 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi vesicle budding(GO:0048194) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.5 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.0 | 0.2 | GO:0007128 | meiotic prophase I(GO:0007128) |
| 0.0 | 0.1 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.1 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.0 | 0.1 | GO:0006379 | mRNA cleavage(GO:0006379) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 0.4 | GO:0004083 | bisphosphoglycerate mutase activity(GO:0004082) bisphosphoglycerate 2-phosphatase activity(GO:0004083) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.3 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.8 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.0 | 0.3 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.2 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.2 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.2 | GO:0005537 | mannose binding(GO:0005537) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | ST_ERK1_ERK2_MAPK_PATHWAY | ERK1/ERK2 MAPK Pathway |