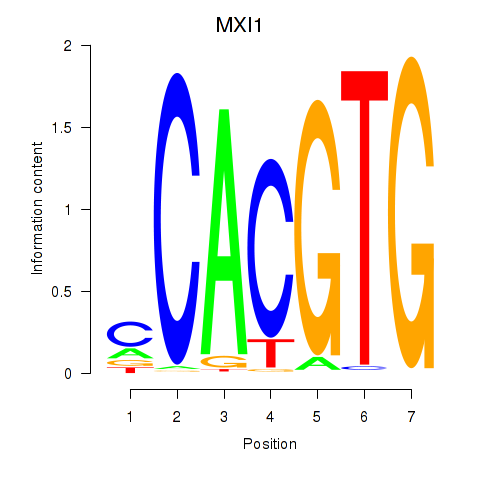
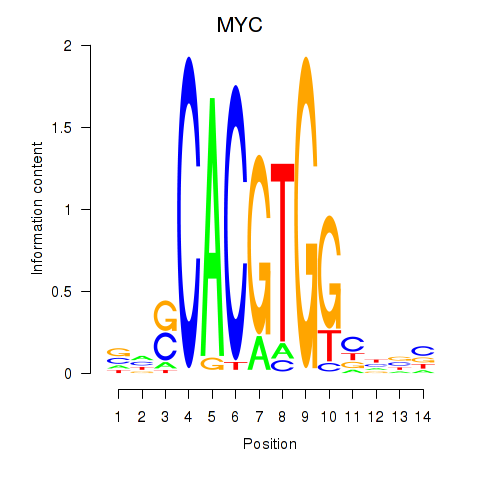
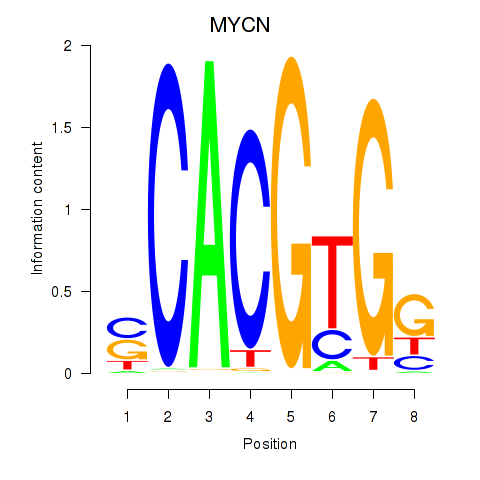
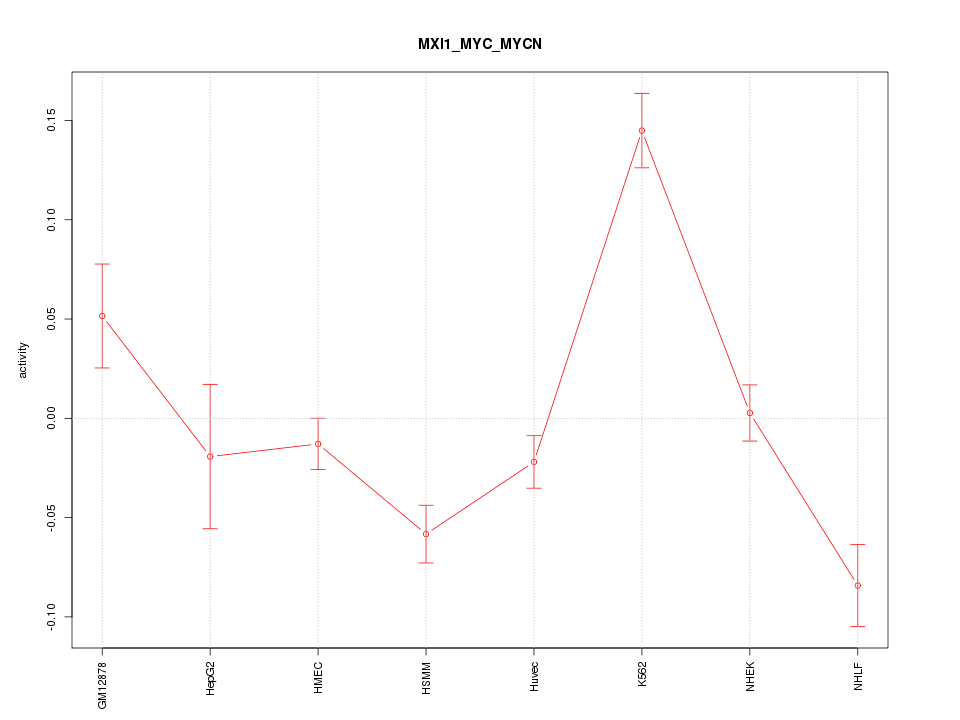
| 2.1 |
6.4 |
GO:0035408 |
histone H3-T6 phosphorylation(GO:0035408) |
| 2.0 |
6.0 |
GO:0033025 |
mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 1.7 |
13.7 |
GO:0006188 |
IMP biosynthetic process(GO:0006188) IMP metabolic process(GO:0046040) |
| 1.7 |
5.1 |
GO:0000738 |
DNA catabolic process, exonucleolytic(GO:0000738) |
| 1.7 |
5.0 |
GO:0070407 |
oxidation-dependent protein catabolic process(GO:0070407) |
| 1.5 |
8.8 |
GO:0060397 |
JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 1.4 |
4.1 |
GO:0042760 |
very long-chain fatty acid catabolic process(GO:0042760) |
| 1.4 |
5.5 |
GO:0051534 |
negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 1.4 |
4.1 |
GO:0046604 |
regulation of mitotic centrosome separation(GO:0046602) positive regulation of mitotic centrosome separation(GO:0046604) |
| 1.3 |
5.2 |
GO:0061032 |
visceral serous pericardium development(GO:0061032) |
| 1.3 |
6.5 |
GO:0070813 |
cysteine biosynthetic process(GO:0019344) hydrogen sulfide metabolic process(GO:0070813) hydrogen sulfide biosynthetic process(GO:0070814) |
| 1.2 |
4.9 |
GO:0010944 |
negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 1.2 |
8.5 |
GO:0015671 |
oxygen transport(GO:0015671) |
| 1.2 |
7.1 |
GO:0043985 |
histone H4-R3 methylation(GO:0043985) |
| 1.2 |
3.5 |
GO:0006273 |
lagging strand elongation(GO:0006273) |
| 1.1 |
8.8 |
GO:0045039 |
protein import into mitochondrial inner membrane(GO:0045039) |
| 1.0 |
10.3 |
GO:0009113 |
purine nucleobase biosynthetic process(GO:0009113) |
| 1.0 |
5.1 |
GO:0007144 |
female meiosis I(GO:0007144) |
| 1.0 |
1.0 |
GO:0046102 |
inosine metabolic process(GO:0046102) |
| 1.0 |
4.0 |
GO:0019348 |
polyprenol metabolic process(GO:0016093) dolichol metabolic process(GO:0019348) |
| 0.9 |
2.6 |
GO:0034626 |
fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.9 |
4.4 |
GO:0008295 |
spermidine biosynthetic process(GO:0008295) |
| 0.9 |
3.5 |
GO:0032929 |
negative regulation of superoxide anion generation(GO:0032929) |
| 0.9 |
4.3 |
GO:0010508 |
positive regulation of autophagy(GO:0010508) |
| 0.8 |
3.4 |
GO:0019264 |
glycine biosynthetic process from serine(GO:0019264) |
| 0.8 |
3.3 |
GO:0046112 |
nucleobase biosynthetic process(GO:0046112) |
| 0.8 |
2.3 |
GO:0031118 |
rRNA pseudouridine synthesis(GO:0031118) |
| 0.7 |
3.0 |
GO:0043490 |
malate-aspartate shuttle(GO:0043490) |
| 0.7 |
6.0 |
GO:0006474 |
N-terminal protein amino acid acetylation(GO:0006474) |
| 0.7 |
2.2 |
GO:0045618 |
positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.7 |
4.4 |
GO:0033160 |
positive regulation of protein import into nucleus, translocation(GO:0033160) |
| 0.7 |
2.9 |
GO:0000463 |
maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) maturation of LSU-rRNA(GO:0000470) |
| 0.7 |
2.1 |
GO:1990170 |
detoxification of cadmium ion(GO:0071585) stress response to cadmium ion(GO:1990170) |
| 0.7 |
3.6 |
GO:0060528 |
secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.7 |
4.3 |
GO:0006552 |
leucine catabolic process(GO:0006552) |
| 0.7 |
2.1 |
GO:0070734 |
histone H3-K27 methylation(GO:0070734) |
| 0.7 |
3.4 |
GO:0010447 |
response to acidic pH(GO:0010447) |
| 0.7 |
3.3 |
GO:0032959 |
inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.6 |
5.0 |
GO:0045008 |
depyrimidination(GO:0045008) |
| 0.6 |
1.9 |
GO:0070171 |
negative regulation of tooth mineralization(GO:0070171) |
| 0.6 |
3.7 |
GO:0046541 |
saliva secretion(GO:0046541) |
| 0.6 |
2.4 |
GO:0000394 |
RNA splicing, via endonucleolytic cleavage and ligation(GO:0000394) tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.6 |
3.6 |
GO:1901984 |
negative regulation of histone acetylation(GO:0035067) negative regulation of protein acetylation(GO:1901984) negative regulation of peptidyl-lysine acetylation(GO:2000757) |
| 0.6 |
1.2 |
GO:0032025 |
response to cobalt ion(GO:0032025) |
| 0.6 |
2.3 |
GO:0006384 |
transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.6 |
2.3 |
GO:0010157 |
response to chlorate(GO:0010157) |
| 0.6 |
5.1 |
GO:0006546 |
glycine catabolic process(GO:0006546) |
| 0.6 |
1.7 |
GO:0019242 |
methylglyoxal biosynthetic process(GO:0019242) |
| 0.5 |
5.4 |
GO:0001522 |
pseudouridine synthesis(GO:0001522) |
| 0.5 |
3.3 |
GO:0060766 |
negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.5 |
2.7 |
GO:0070935 |
3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.5 |
1.6 |
GO:0019695 |
choline metabolic process(GO:0019695) |
| 0.5 |
5.0 |
GO:0042535 |
positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.5 |
4.5 |
GO:0070327 |
thyroid hormone transport(GO:0070327) |
| 0.5 |
3.0 |
GO:0000076 |
DNA replication checkpoint(GO:0000076) |
| 0.5 |
1.5 |
GO:0061198 |
fungiform papilla development(GO:0061196) fungiform papilla morphogenesis(GO:0061197) fungiform papilla formation(GO:0061198) |
| 0.5 |
7.0 |
GO:0006596 |
polyamine biosynthetic process(GO:0006596) |
| 0.5 |
3.9 |
GO:0015862 |
uridine transport(GO:0015862) |
| 0.5 |
7.3 |
GO:0031115 |
negative regulation of microtubule polymerization(GO:0031115) |
| 0.5 |
2.9 |
GO:0035751 |
lysosomal lumen acidification(GO:0007042) regulation of lysosomal lumen pH(GO:0035751) |
| 0.5 |
1.4 |
GO:0048478 |
replication fork protection(GO:0048478) |
| 0.5 |
2.4 |
GO:0032203 |
telomere formation via telomerase(GO:0032203) |
| 0.5 |
1.4 |
GO:0006432 |
phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.5 |
6.0 |
GO:0006386 |
transcription elongation from RNA polymerase III promoter(GO:0006385) termination of RNA polymerase III transcription(GO:0006386) |
| 0.5 |
1.4 |
GO:0001833 |
inner cell mass cell proliferation(GO:0001833) |
| 0.5 |
2.7 |
GO:0006999 |
nuclear pore organization(GO:0006999) |
| 0.4 |
4.0 |
GO:0032464 |
positive regulation of protein homooligomerization(GO:0032464) |
| 0.4 |
8.2 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.4 |
0.9 |
GO:0072384 |
organelle transport along microtubule(GO:0072384) |
| 0.4 |
10.3 |
GO:0000291 |
nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.4 |
3.4 |
GO:0007090 |
obsolete regulation of S phase of mitotic cell cycle(GO:0007090) |
| 0.4 |
1.7 |
GO:0000966 |
RNA 5'-end processing(GO:0000966) |
| 0.4 |
9.4 |
GO:0071577 |
zinc II ion transmembrane transport(GO:0071577) |
| 0.4 |
0.8 |
GO:0032608 |
interferon-beta production(GO:0032608) regulation of interferon-beta production(GO:0032648) |
| 0.4 |
0.8 |
GO:0015920 |
lipopolysaccharide transport(GO:0015920) |
| 0.4 |
2.0 |
GO:1901798 |
positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.4 |
2.0 |
GO:0007143 |
female meiotic division(GO:0007143) |
| 0.4 |
1.2 |
GO:0070445 |
Fc receptor mediated stimulatory signaling pathway(GO:0002431) regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.4 |
6.3 |
GO:0007253 |
cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.4 |
2.4 |
GO:0009082 |
branched-chain amino acid biosynthetic process(GO:0009082) |
| 0.4 |
1.2 |
GO:0014022 |
neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.4 |
0.8 |
GO:0060087 |
relaxation of smooth muscle(GO:0044557) relaxation of vascular smooth muscle(GO:0060087) |
| 0.4 |
2.6 |
GO:0015755 |
fructose transport(GO:0015755) |
| 0.4 |
1.8 |
GO:0035455 |
response to interferon-alpha(GO:0035455) |
| 0.4 |
5.4 |
GO:0006400 |
tRNA modification(GO:0006400) |
| 0.4 |
0.7 |
GO:0006166 |
purine ribonucleoside salvage(GO:0006166) dATP metabolic process(GO:0046060) |
| 0.4 |
1.1 |
GO:0071344 |
diphosphate metabolic process(GO:0071344) |
| 0.3 |
2.1 |
GO:0006422 |
aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.3 |
3.8 |
GO:0051085 |
chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.3 |
1.7 |
GO:0009143 |
nucleoside triphosphate catabolic process(GO:0009143) |
| 0.3 |
0.7 |
GO:0006419 |
alanyl-tRNA aminoacylation(GO:0006419) |
| 0.3 |
1.6 |
GO:0071243 |
cellular response to arsenic-containing substance(GO:0071243) |
| 0.3 |
1.3 |
GO:0051503 |
adenine nucleotide transport(GO:0051503) |
| 0.3 |
1.0 |
GO:0010586 |
miRNA metabolic process(GO:0010586) miRNA catabolic process(GO:0010587) |
| 0.3 |
1.6 |
GO:0003093 |
regulation of glomerular filtration(GO:0003093) glomerular filtration(GO:0003094) renal filtration(GO:0097205) regulation of renal system process(GO:0098801) |
| 0.3 |
0.3 |
GO:0006405 |
RNA export from nucleus(GO:0006405) |
| 0.3 |
0.6 |
GO:0006101 |
citrate metabolic process(GO:0006101) |
| 0.3 |
0.9 |
GO:0031034 |
skeletal muscle myosin thick filament assembly(GO:0030241) myosin filament assembly(GO:0031034) striated muscle myosin thick filament assembly(GO:0071688) |
| 0.3 |
0.9 |
GO:0006344 |
maintenance of chromatin silencing(GO:0006344) |
| 0.3 |
1.5 |
GO:0017055 |
negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.3 |
0.9 |
GO:0019858 |
cytosine metabolic process(GO:0019858) |
| 0.3 |
6.7 |
GO:0015701 |
bicarbonate transport(GO:0015701) |
| 0.3 |
10.4 |
GO:0006270 |
DNA replication initiation(GO:0006270) |
| 0.3 |
1.1 |
GO:0006391 |
transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.3 |
2.3 |
GO:0006452 |
translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.3 |
2.6 |
GO:0048387 |
negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.3 |
1.1 |
GO:0016074 |
snoRNA metabolic process(GO:0016074) |
| 0.3 |
1.7 |
GO:0045654 |
positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.3 |
1.1 |
GO:0042816 |
pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.3 |
1.7 |
GO:0018216 |
peptidyl-arginine methylation(GO:0018216) |
| 0.3 |
8.1 |
GO:0006362 |
transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.3 |
1.1 |
GO:0045875 |
negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.3 |
1.1 |
GO:0006089 |
lactate metabolic process(GO:0006089) |
| 0.3 |
1.1 |
GO:0048935 |
peripheral nervous system neuron development(GO:0048935) |
| 0.3 |
2.4 |
GO:0006577 |
amino-acid betaine metabolic process(GO:0006577) carnitine metabolic process(GO:0009437) |
| 0.3 |
1.1 |
GO:0000491 |
small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.3 |
1.0 |
GO:0048642 |
negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.3 |
0.8 |
GO:0009134 |
nucleoside diphosphate catabolic process(GO:0009134) ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.3 |
7.8 |
GO:0006099 |
tricarboxylic acid cycle(GO:0006099) |
| 0.3 |
2.1 |
GO:0046174 |
polyol catabolic process(GO:0046174) |
| 0.3 |
0.8 |
GO:0006325 |
chromatin organization(GO:0006325) |
| 0.3 |
0.3 |
GO:0045176 |
apical protein localization(GO:0045176) |
| 0.3 |
1.0 |
GO:0060748 |
tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.3 |
1.5 |
GO:0006658 |
phosphatidylserine metabolic process(GO:0006658) |
| 0.2 |
1.2 |
GO:0035025 |
positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.2 |
4.7 |
GO:0019674 |
NAD metabolic process(GO:0019674) |
| 0.2 |
1.0 |
GO:0042483 |
negative regulation of odontogenesis(GO:0042483) |
| 0.2 |
0.2 |
GO:0015959 |
diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.2 |
0.7 |
GO:0090073 |
positive regulation of protein homodimerization activity(GO:0090073) |
| 0.2 |
0.7 |
GO:0034551 |
respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.2 |
2.4 |
GO:0051001 |
negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.2 |
0.7 |
GO:0043095 |
regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.2 |
1.4 |
GO:0002478 |
antigen processing and presentation of exogenous peptide antigen(GO:0002478) |
| 0.2 |
0.7 |
GO:0051083 |
'de novo' cotranslational protein folding(GO:0051083) |
| 0.2 |
0.5 |
GO:0072136 |
kidney mesenchymal cell proliferation(GO:0072135) metanephric mesenchymal cell proliferation involved in metanephros development(GO:0072136) |
| 0.2 |
0.7 |
GO:0034086 |
maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.2 |
3.1 |
GO:0002566 |
somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.2 |
0.9 |
GO:0006177 |
GMP biosynthetic process(GO:0006177) |
| 0.2 |
3.3 |
GO:1901663 |
quinone biosynthetic process(GO:1901663) |
| 0.2 |
1.4 |
GO:0022614 |
membrane to membrane docking(GO:0022614) |
| 0.2 |
1.8 |
GO:0015802 |
basic amino acid transport(GO:0015802) |
| 0.2 |
0.7 |
GO:0032196 |
transposition, DNA-mediated(GO:0006313) transposition(GO:0032196) |
| 0.2 |
1.1 |
GO:0017183 |
peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.2 |
28.0 |
GO:0042254 |
ribosome biogenesis(GO:0042254) |
| 0.2 |
1.1 |
GO:0046831 |
regulation of RNA export from nucleus(GO:0046831) |
| 0.2 |
0.7 |
GO:0072387 |
flavin-containing compound biosynthetic process(GO:0042727) flavin adenine dinucleotide metabolic process(GO:0072387) |
| 0.2 |
7.7 |
GO:0000080 |
mitotic G1 phase(GO:0000080) |
| 0.2 |
1.1 |
GO:0043116 |
negative regulation of vascular permeability(GO:0043116) |
| 0.2 |
0.9 |
GO:0051958 |
methotrexate transport(GO:0051958) |
| 0.2 |
1.3 |
GO:0002726 |
positive regulation of T cell cytokine production(GO:0002726) |
| 0.2 |
0.6 |
GO:0040016 |
embryonic cleavage(GO:0040016) |
| 0.2 |
5.4 |
GO:0015804 |
neutral amino acid transport(GO:0015804) |
| 0.2 |
0.4 |
GO:0008625 |
extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) extrinsic apoptotic signaling pathway(GO:0097191) |
| 0.2 |
0.6 |
GO:0034755 |
iron ion transmembrane transport(GO:0034755) |
| 0.2 |
1.1 |
GO:0006975 |
DNA damage induced protein phosphorylation(GO:0006975) |
| 0.2 |
0.4 |
GO:0045005 |
DNA-dependent DNA replication maintenance of fidelity(GO:0045005) |
| 0.2 |
2.9 |
GO:0071378 |
growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.2 |
1.4 |
GO:0006572 |
tyrosine catabolic process(GO:0006572) |
| 0.2 |
3.7 |
GO:0006098 |
pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.2 |
5.7 |
GO:0016925 |
protein sumoylation(GO:0016925) |
| 0.2 |
1.0 |
GO:0032933 |
response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.2 |
2.8 |
GO:0032703 |
negative regulation of interleukin-2 production(GO:0032703) |
| 0.2 |
5.2 |
GO:0060334 |
regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.2 |
1.2 |
GO:0018026 |
peptidyl-lysine monomethylation(GO:0018026) |
| 0.2 |
3.0 |
GO:0031648 |
protein destabilization(GO:0031648) |
| 0.2 |
0.6 |
GO:0034720 |
histone H3-K4 demethylation(GO:0034720) |
| 0.2 |
4.5 |
GO:0031055 |
DNA replication-independent nucleosome assembly(GO:0006336) chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) DNA replication-independent nucleosome organization(GO:0034724) CENP-A containing chromatin organization(GO:0061641) |
| 0.2 |
1.2 |
GO:0006449 |
regulation of translational termination(GO:0006449) |
| 0.2 |
0.4 |
GO:0051293 |
establishment of spindle localization(GO:0051293) spindle localization(GO:0051653) |
| 0.2 |
1.3 |
GO:0032713 |
negative regulation of interleukin-4 production(GO:0032713) |
| 0.2 |
1.0 |
GO:0006564 |
L-serine biosynthetic process(GO:0006564) |
| 0.2 |
0.6 |
GO:0007206 |
phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.2 |
1.1 |
GO:0009128 |
AMP catabolic process(GO:0006196) purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.2 |
0.6 |
GO:0000019 |
regulation of mitotic recombination(GO:0000019) |
| 0.2 |
0.2 |
GO:0001836 |
release of cytochrome c from mitochondria(GO:0001836) |
| 0.2 |
0.7 |
GO:0060013 |
righting reflex(GO:0060013) |
| 0.2 |
1.3 |
GO:0006346 |
methylation-dependent chromatin silencing(GO:0006346) |
| 0.2 |
0.2 |
GO:0035330 |
regulation of hippo signaling(GO:0035330) |
| 0.2 |
1.1 |
GO:0071699 |
olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.2 |
0.7 |
GO:0072106 |
regulation of ureteric bud formation(GO:0072106) positive regulation of ureteric bud formation(GO:0072107) |
| 0.2 |
0.5 |
GO:0046958 |
operant conditioning(GO:0035106) nonassociative learning(GO:0046958) |
| 0.2 |
0.9 |
GO:0045767 |
obsolete regulation of anti-apoptosis(GO:0045767) |
| 0.2 |
0.5 |
GO:0046080 |
dUTP metabolic process(GO:0046080) |
| 0.2 |
0.4 |
GO:0060603 |
mammary gland duct morphogenesis(GO:0060603) |
| 0.2 |
6.7 |
GO:0008033 |
tRNA processing(GO:0008033) |
| 0.2 |
0.4 |
GO:0045719 |
negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.2 |
0.9 |
GO:0019509 |
L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.2 |
1.4 |
GO:0006020 |
inositol metabolic process(GO:0006020) |
| 0.2 |
1.6 |
GO:0032508 |
DNA duplex unwinding(GO:0032508) |
| 0.2 |
1.4 |
GO:0016576 |
histone dephosphorylation(GO:0016576) |
| 0.2 |
0.3 |
GO:1903301 |
regulation of glucokinase activity(GO:0033131) positive regulation of glucokinase activity(GO:0033133) regulation of hexokinase activity(GO:1903299) positive regulation of hexokinase activity(GO:1903301) |
| 0.2 |
0.9 |
GO:0046037 |
GMP metabolic process(GO:0046037) |
| 0.2 |
1.0 |
GO:0019372 |
lipoxygenase pathway(GO:0019372) |
| 0.2 |
0.3 |
GO:0048311 |
mitochondrion distribution(GO:0048311) |
| 0.2 |
0.5 |
GO:0071104 |
negative regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0003340) response to interleukin-9(GO:0071104) response to interleukin-11(GO:0071105) |
| 0.2 |
2.5 |
GO:0051131 |
chaperone-mediated protein complex assembly(GO:0051131) |
| 0.2 |
1.2 |
GO:0045007 |
base-excision repair, AP site formation(GO:0006285) depurination(GO:0045007) |
| 0.2 |
0.7 |
GO:0051013 |
microtubule severing(GO:0051013) |
| 0.2 |
1.2 |
GO:0016265 |
obsolete death(GO:0016265) |
| 0.2 |
1.3 |
GO:0006561 |
proline biosynthetic process(GO:0006561) |
| 0.2 |
0.2 |
GO:0009893 |
positive regulation of metabolic process(GO:0009893) |
| 0.2 |
0.7 |
GO:0048388 |
endosomal lumen acidification(GO:0048388) |
| 0.2 |
0.7 |
GO:0006668 |
sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.2 |
0.3 |
GO:0035405 |
histone-threonine phosphorylation(GO:0035405) |
| 0.2 |
0.6 |
GO:0000012 |
single strand break repair(GO:0000012) |
| 0.2 |
0.6 |
GO:0030259 |
lipid glycosylation(GO:0030259) |
| 0.2 |
0.2 |
GO:0033002 |
muscle cell proliferation(GO:0033002) |
| 0.2 |
0.3 |
GO:0030578 |
nuclear body organization(GO:0030575) PML body organization(GO:0030578) |
| 0.2 |
2.6 |
GO:0071451 |
removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.2 |
1.3 |
GO:0010038 |
response to metal ion(GO:0010038) |
| 0.2 |
0.5 |
GO:0060259 |
regulation of feeding behavior(GO:0060259) |
| 0.2 |
0.5 |
GO:0071539 |
Golgi localization(GO:0051645) protein localization to centrosome(GO:0071539) |
| 0.2 |
0.8 |
GO:0006686 |
sphingomyelin biosynthetic process(GO:0006686) |
| 0.2 |
0.5 |
GO:0032957 |
inositol trisphosphate metabolic process(GO:0032957) |
| 0.2 |
0.5 |
GO:0052646 |
glycerol-3-phosphate metabolic process(GO:0006072) alditol phosphate metabolic process(GO:0052646) |
| 0.2 |
0.9 |
GO:0043984 |
histone H4-K16 acetylation(GO:0043984) |
| 0.2 |
1.4 |
GO:1901070 |
guanosine-containing compound biosynthetic process(GO:1901070) |
| 0.2 |
2.0 |
GO:0097031 |
NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.2 |
0.5 |
GO:0034498 |
early endosome to Golgi transport(GO:0034498) |
| 0.2 |
2.3 |
GO:0045742 |
positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) positive regulation of ERBB signaling pathway(GO:1901186) |
| 0.1 |
1.5 |
GO:0033261 |
obsolete regulation of S phase(GO:0033261) |
| 0.1 |
0.4 |
GO:1901522 |
positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.1 |
0.3 |
GO:0031016 |
pancreas development(GO:0031016) |
| 0.1 |
0.6 |
GO:0006408 |
snRNA export from nucleus(GO:0006408) snRNA transport(GO:0051030) |
| 0.1 |
2.3 |
GO:0019221 |
cytokine-mediated signaling pathway(GO:0019221) |
| 0.1 |
7.9 |
GO:0006369 |
termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 |
0.4 |
GO:0001732 |
formation of cytoplasmic translation initiation complex(GO:0001732) cytoplasmic translation(GO:0002181) cytoplasmic translational initiation(GO:0002183) |
| 0.1 |
0.8 |
GO:0000303 |
response to superoxide(GO:0000303) |
| 0.1 |
0.3 |
GO:0032898 |
neurotrophin production(GO:0032898) |
| 0.1 |
0.4 |
GO:0010896 |
regulation of triglyceride catabolic process(GO:0010896) positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.1 |
0.8 |
GO:0045084 |
positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.1 |
0.4 |
GO:0006782 |
protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.1 |
0.4 |
GO:0042759 |
long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 |
0.1 |
GO:0043551 |
regulation of phosphatidylinositol 3-kinase activity(GO:0043551) positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.1 |
0.4 |
GO:0021529 |
spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) glial cell fate specification(GO:0021780) |
| 0.1 |
1.3 |
GO:0015937 |
coenzyme A biosynthetic process(GO:0015937) |
| 0.1 |
0.1 |
GO:0040019 |
positive regulation of embryonic development(GO:0040019) |
| 0.1 |
0.4 |
GO:0009070 |
serine family amino acid biosynthetic process(GO:0009070) |
| 0.1 |
0.5 |
GO:0046386 |
pyrimidine nucleotide catabolic process(GO:0006244) pyrimidine deoxyribonucleotide catabolic process(GO:0009223) deoxyribose phosphate catabolic process(GO:0046386) |
| 0.1 |
1.0 |
GO:0015074 |
DNA integration(GO:0015074) |
| 0.1 |
0.5 |
GO:0019724 |
B cell mediated immunity(GO:0019724) |
| 0.1 |
0.8 |
GO:0060707 |
trophoblast giant cell differentiation(GO:0060707) |
| 0.1 |
0.8 |
GO:0007320 |
insemination(GO:0007320) |
| 0.1 |
1.0 |
GO:0009396 |
folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.1 |
1.8 |
GO:0048261 |
negative regulation of receptor-mediated endocytosis(GO:0048261) |
| 0.1 |
0.5 |
GO:0006553 |
lysine metabolic process(GO:0006553) lysine catabolic process(GO:0006554) |
| 0.1 |
5.2 |
GO:0043038 |
tRNA aminoacylation for protein translation(GO:0006418) amino acid activation(GO:0043038) tRNA aminoacylation(GO:0043039) |
| 0.1 |
3.6 |
GO:0071804 |
cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.1 |
0.5 |
GO:0001550 |
ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 |
1.6 |
GO:0030388 |
fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 |
0.5 |
GO:0006498 |
N-terminal protein lipidation(GO:0006498) |
| 0.1 |
0.5 |
GO:0010826 |
negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.1 |
0.6 |
GO:0032878 |
regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.1 |
1.0 |
GO:0043084 |
penile erection(GO:0043084) |
| 0.1 |
0.4 |
GO:0032927 |
positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.1 |
3.9 |
GO:0006626 |
protein targeting to mitochondrion(GO:0006626) |
| 0.1 |
0.8 |
GO:0032375 |
negative regulation of sterol transport(GO:0032372) negative regulation of cholesterol transport(GO:0032375) |
| 0.1 |
1.4 |
GO:0009303 |
rRNA transcription(GO:0009303) |
| 0.1 |
3.4 |
GO:0036503 |
ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) ERAD pathway(GO:0036503) |
| 0.1 |
0.9 |
GO:0017004 |
cytochrome complex assembly(GO:0017004) |
| 0.1 |
1.5 |
GO:0006839 |
mitochondrial transport(GO:0006839) |
| 0.1 |
1.0 |
GO:0046835 |
carbohydrate phosphorylation(GO:0046835) |
| 0.1 |
0.8 |
GO:0033962 |
cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.1 |
1.7 |
GO:0030641 |
regulation of cellular pH(GO:0030641) |
| 0.1 |
2.6 |
GO:0042776 |
mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 |
0.3 |
GO:0033364 |
endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) mast cell secretory granule organization(GO:0033364) |
| 0.1 |
0.1 |
GO:2000059 |
negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000059) |
| 0.1 |
0.1 |
GO:0090342 |
regulation of cell aging(GO:0090342) |
| 0.1 |
0.2 |
GO:0060080 |
inhibitory postsynaptic potential(GO:0060080) |
| 0.1 |
0.3 |
GO:0018916 |
nitrobenzene metabolic process(GO:0018916) |
| 0.1 |
0.2 |
GO:0042262 |
DNA protection(GO:0042262) |
| 0.1 |
0.2 |
GO:0032355 |
response to estradiol(GO:0032355) |
| 0.1 |
1.2 |
GO:0046902 |
regulation of mitochondrial membrane permeability(GO:0046902) regulation of membrane permeability(GO:0090559) |
| 0.1 |
0.7 |
GO:0043457 |
regulation of cellular respiration(GO:0043457) |
| 0.1 |
0.3 |
GO:0046320 |
regulation of fatty acid oxidation(GO:0046320) |
| 0.1 |
0.3 |
GO:0090267 |
positive regulation of spindle checkpoint(GO:0090232) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) positive regulation of cell cycle checkpoint(GO:1901978) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.1 |
0.8 |
GO:0046655 |
folic acid metabolic process(GO:0046655) |
| 0.1 |
1.1 |
GO:0006776 |
vitamin A metabolic process(GO:0006776) |
| 0.1 |
0.7 |
GO:0003310 |
pancreatic A cell differentiation(GO:0003310) |
| 0.1 |
0.3 |
GO:0021978 |
telencephalon regionalization(GO:0021978) |
| 0.1 |
2.2 |
GO:0043496 |
regulation of protein homodimerization activity(GO:0043496) |
| 0.1 |
0.4 |
GO:0071894 |
histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.1 |
0.3 |
GO:0006929 |
substrate-dependent cell migration(GO:0006929) |
| 0.1 |
0.5 |
GO:0006000 |
fructose metabolic process(GO:0006000) |
| 0.1 |
0.5 |
GO:0042415 |
norepinephrine metabolic process(GO:0042415) |
| 0.1 |
0.4 |
GO:0001831 |
trophectodermal cellular morphogenesis(GO:0001831) |
| 0.1 |
0.4 |
GO:0031580 |
membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 |
4.2 |
GO:0008156 |
negative regulation of DNA replication(GO:0008156) |
| 0.1 |
11.6 |
GO:0000279 |
M phase(GO:0000279) |
| 0.1 |
0.1 |
GO:0042045 |
epithelial fluid transport(GO:0042045) |
| 0.1 |
0.5 |
GO:0031664 |
regulation of lipopolysaccharide-mediated signaling pathway(GO:0031664) negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.1 |
0.4 |
GO:0072207 |
metanephric tubule development(GO:0072170) metanephric epithelium development(GO:0072207) metanephric nephron tubule development(GO:0072234) metanephric nephron epithelium development(GO:0072243) |
| 0.1 |
0.2 |
GO:0052556 |
pathogen-associated molecular pattern dependent induction by symbiont of host innate immune response(GO:0052033) positive regulation by symbiont of host innate immune response(GO:0052166) modulation by symbiont of host innate immune response(GO:0052167) pathogen-associated molecular pattern dependent modulation by symbiont of host innate immune response(GO:0052169) pathogen-associated molecular pattern dependent induction by organism of innate immune response of other organism involved in symbiotic interaction(GO:0052257) positive regulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052305) modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052306) pathogen-associated molecular pattern dependent modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052308) positive regulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052555) positive regulation by symbiont of host immune response(GO:0052556) |
| 0.1 |
0.1 |
GO:0003056 |
regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.1 |
0.7 |
GO:0051103 |
DNA ligation involved in DNA repair(GO:0051103) |
| 0.1 |
0.2 |
GO:0008633 |
obsolete activation of pro-apoptotic gene products(GO:0008633) |
| 0.1 |
1.2 |
GO:0006582 |
melanin metabolic process(GO:0006582) |
| 0.1 |
2.8 |
GO:0006611 |
protein export from nucleus(GO:0006611) |
| 0.1 |
0.5 |
GO:0006703 |
estrogen biosynthetic process(GO:0006703) |
| 0.1 |
3.5 |
GO:0043966 |
histone H3 acetylation(GO:0043966) |
| 0.1 |
0.6 |
GO:0006855 |
drug transmembrane transport(GO:0006855) |
| 0.1 |
0.3 |
GO:0070384 |
Harderian gland development(GO:0070384) |
| 0.1 |
1.2 |
GO:0008635 |
activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 |
0.2 |
GO:0032223 |
negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.1 |
0.2 |
GO:0060120 |
auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.1 |
0.2 |
GO:0002378 |
immunoglobulin biosynthetic process(GO:0002378) |
| 0.1 |
0.4 |
GO:0015886 |
heme transport(GO:0015886) |
| 0.1 |
2.4 |
GO:0006904 |
vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 |
2.5 |
GO:0033572 |
ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.1 |
0.5 |
GO:0016561 |
protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.1 |
0.7 |
GO:0016180 |
snRNA processing(GO:0016180) |
| 0.1 |
0.5 |
GO:0009649 |
entrainment of circadian clock(GO:0009649) |
| 0.1 |
0.2 |
GO:0070781 |
response to biotin(GO:0070781) |
| 0.1 |
0.6 |
GO:0045773 |
positive regulation of axon extension(GO:0045773) |
| 0.1 |
1.0 |
GO:0006376 |
mRNA splice site selection(GO:0006376) |
| 0.1 |
1.2 |
GO:0032008 |
positive regulation of TOR signaling(GO:0032008) |
| 0.1 |
0.1 |
GO:0033689 |
negative regulation of osteoblast proliferation(GO:0033689) |
| 0.1 |
0.5 |
GO:0006885 |
regulation of pH(GO:0006885) |
| 0.1 |
5.5 |
GO:0006968 |
cellular defense response(GO:0006968) |
| 0.1 |
0.2 |
GO:0006041 |
glucosamine metabolic process(GO:0006041) |
| 0.1 |
0.3 |
GO:0034723 |
DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 |
0.7 |
GO:0048712 |
negative regulation of astrocyte differentiation(GO:0048712) |
| 0.1 |
0.3 |
GO:0046952 |
ketone body catabolic process(GO:0046952) |
| 0.1 |
1.1 |
GO:0001662 |
behavioral fear response(GO:0001662) |
| 0.1 |
0.5 |
GO:0045987 |
positive regulation of smooth muscle contraction(GO:0045987) |
| 0.1 |
1.1 |
GO:0006665 |
sphingolipid metabolic process(GO:0006665) |
| 0.1 |
0.2 |
GO:0034635 |
glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.1 |
0.3 |
GO:0051964 |
negative regulation of synapse assembly(GO:0051964) |
| 0.1 |
0.4 |
GO:0018343 |
protein farnesylation(GO:0018343) |
| 0.1 |
0.1 |
GO:0032060 |
bleb assembly(GO:0032060) regulation of cytolysis(GO:0042268) pore complex assembly(GO:0046931) |
| 0.1 |
0.1 |
GO:0002035 |
brain renin-angiotensin system(GO:0002035) |
| 0.1 |
0.4 |
GO:0071044 |
histone mRNA catabolic process(GO:0071044) |
| 0.1 |
0.4 |
GO:0009120 |
deoxyribonucleoside metabolic process(GO:0009120) |
| 0.1 |
0.1 |
GO:0002577 |
dendritic cell antigen processing and presentation(GO:0002468) regulation of antigen processing and presentation(GO:0002577) positive regulation of antigen processing and presentation(GO:0002579) regulation of dendritic cell antigen processing and presentation(GO:0002604) positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.1 |
0.9 |
GO:1904031 |
positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.1 |
0.3 |
GO:0060749 |
mammary gland alveolus development(GO:0060749) mammary gland lobule development(GO:0061377) |
| 0.1 |
0.2 |
GO:0000114 |
obsolete regulation of transcription involved in G1 phase of mitotic cell cycle(GO:0000114) |
| 0.1 |
0.1 |
GO:0030948 |
negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.1 |
0.4 |
GO:0008089 |
anterograde axonal transport(GO:0008089) |
| 0.1 |
0.3 |
GO:0045604 |
regulation of epidermal cell differentiation(GO:0045604) |
| 0.1 |
0.4 |
GO:0051567 |
histone H3-K9 methylation(GO:0051567) histone H3-K9 modification(GO:0061647) |
| 0.1 |
0.4 |
GO:0051258 |
protein polymerization(GO:0051258) |
| 0.1 |
0.4 |
GO:0000729 |
DNA double-strand break processing(GO:0000729) |
| 0.1 |
0.2 |
GO:0045945 |
positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.1 |
0.2 |
GO:1901998 |
tetracycline transport(GO:0015904) antibiotic transport(GO:0042891) toxin transport(GO:1901998) |
| 0.1 |
2.2 |
GO:0060041 |
retina development in camera-type eye(GO:0060041) |
| 0.1 |
1.2 |
GO:0010467 |
gene expression(GO:0010467) |
| 0.1 |
0.9 |
GO:0000910 |
cytokinesis(GO:0000910) |
| 0.1 |
1.0 |
GO:0033014 |
porphyrin-containing compound biosynthetic process(GO:0006779) tetrapyrrole biosynthetic process(GO:0033014) |
| 0.1 |
0.4 |
GO:0035058 |
nonmotile primary cilium assembly(GO:0035058) |
| 0.1 |
0.1 |
GO:0045347 |
negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.1 |
0.4 |
GO:0070167 |
regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.1 |
1.8 |
GO:0070830 |
bicellular tight junction assembly(GO:0070830) |
| 0.1 |
0.6 |
GO:0048854 |
brain morphogenesis(GO:0048854) |
| 0.1 |
0.3 |
GO:0008228 |
opsonization(GO:0008228) |
| 0.1 |
1.2 |
GO:0006111 |
regulation of gluconeogenesis(GO:0006111) |
| 0.1 |
0.4 |
GO:0007021 |
tubulin complex assembly(GO:0007021) |
| 0.1 |
0.4 |
GO:0002360 |
T cell lineage commitment(GO:0002360) |
| 0.1 |
0.2 |
GO:0032486 |
Rap protein signal transduction(GO:0032486) |
| 0.1 |
0.7 |
GO:0050858 |
negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 |
2.8 |
GO:0007612 |
learning(GO:0007612) |
| 0.1 |
0.2 |
GO:0007207 |
phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 |
0.2 |
GO:0030644 |
cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.1 |
0.2 |
GO:0032506 |
cytokinetic process(GO:0032506) |
| 0.1 |
0.2 |
GO:0006890 |
retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 |
0.1 |
GO:0001306 |
age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.1 |
0.5 |
GO:0006614 |
SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 |
0.2 |
GO:0006002 |
fructose 6-phosphate metabolic process(GO:0006002) |
| 0.1 |
0.2 |
GO:0003149 |
membranous septum morphogenesis(GO:0003149) |
| 0.1 |
0.4 |
GO:0006613 |
cotranslational protein targeting to membrane(GO:0006613) |
| 0.1 |
0.5 |
GO:0006600 |
creatine metabolic process(GO:0006600) |
| 0.1 |
1.4 |
GO:0006699 |
bile acid biosynthetic process(GO:0006699) |
| 0.1 |
0.5 |
GO:0060762 |
regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.1 |
2.6 |
GO:0051084 |
'de novo' posttranslational protein folding(GO:0051084) |
| 0.1 |
1.1 |
GO:0045814 |
negative regulation of gene expression, epigenetic(GO:0045814) |
| 0.1 |
0.3 |
GO:0001880 |
Mullerian duct regression(GO:0001880) |
| 0.1 |
4.9 |
GO:0051028 |
mRNA transport(GO:0051028) |
| 0.1 |
0.2 |
GO:0032364 |
oxygen homeostasis(GO:0032364) |
| 0.1 |
5.0 |
GO:0030041 |
actin filament polymerization(GO:0030041) |
| 0.1 |
0.9 |
GO:0043124 |
negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.1 |
0.5 |
GO:0031295 |
lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.1 |
0.2 |
GO:0046586 |
regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.1 |
0.2 |
GO:0043249 |
erythrocyte maturation(GO:0043249) |
| 0.1 |
0.7 |
GO:0048246 |
macrophage chemotaxis(GO:0048246) |
| 0.1 |
1.0 |
GO:0050853 |
B cell receptor signaling pathway(GO:0050853) |
| 0.1 |
0.5 |
GO:0032092 |
positive regulation of protein binding(GO:0032092) |
| 0.1 |
0.2 |
GO:0070498 |
interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.1 |
0.8 |
GO:0051865 |
protein autoubiquitination(GO:0051865) |
| 0.1 |
1.1 |
GO:0007163 |
establishment or maintenance of cell polarity(GO:0007163) |
| 0.1 |
0.2 |
GO:0001830 |
trophectodermal cell fate commitment(GO:0001830) |
| 0.1 |
0.7 |
GO:0050766 |
positive regulation of phagocytosis(GO:0050766) |
| 0.1 |
1.2 |
GO:0042475 |
odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.1 |
0.2 |
GO:0008612 |
peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 |
0.4 |
GO:0095500 |
G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.0 |
0.1 |
GO:0042560 |
10-formyltetrahydrofolate metabolic process(GO:0009256) 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.0 |
0.1 |
GO:0006857 |
oligopeptide transport(GO:0006857) |
| 0.0 |
0.9 |
GO:0051169 |
nuclear transport(GO:0051169) |
| 0.0 |
0.1 |
GO:0051956 |
negative regulation of organic acid transport(GO:0032891) negative regulation of amino acid transport(GO:0051956) |
| 0.0 |
0.1 |
GO:0043654 |
recognition of apoptotic cell(GO:0043654) |
| 0.0 |
1.0 |
GO:0006607 |
NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 |
9.1 |
GO:0006184 |
obsolete GTP catabolic process(GO:0006184) |
| 0.0 |
1.8 |
GO:0006338 |
chromatin remodeling(GO:0006338) |
| 0.0 |
0.2 |
GO:0021554 |
optic nerve development(GO:0021554) optic nerve morphogenesis(GO:0021631) |
| 0.0 |
0.4 |
GO:0001706 |
endoderm formation(GO:0001706) |
| 0.0 |
0.6 |
GO:0045086 |
positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.0 |
0.1 |
GO:0007389 |
pattern specification process(GO:0007389) |
| 0.0 |
0.0 |
GO:0032392 |
DNA geometric change(GO:0032392) |
| 0.0 |
0.9 |
GO:0016079 |
synaptic vesicle exocytosis(GO:0016079) |
| 0.0 |
0.2 |
GO:0034332 |
adherens junction organization(GO:0034332) |
| 0.0 |
0.1 |
GO:0045065 |
cytotoxic T cell differentiation(GO:0045065) |
| 0.0 |
0.3 |
GO:0015781 |
pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 |
0.2 |
GO:0045730 |
respiratory burst(GO:0045730) |
| 0.0 |
0.2 |
GO:2000273 |
positive regulation of receptor activity(GO:2000273) |
| 0.0 |
0.4 |
GO:0061756 |
leukocyte tethering or rolling(GO:0050901) leukocyte adhesion to vascular endothelial cell(GO:0061756) |
| 0.0 |
0.4 |
GO:0043252 |
sodium-independent organic anion transport(GO:0043252) |
| 0.0 |
1.3 |
GO:0030073 |
insulin secretion(GO:0030073) |
| 0.0 |
0.4 |
GO:0030539 |
male genitalia development(GO:0030539) |
| 0.0 |
0.0 |
GO:0021988 |
olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 |
0.6 |
GO:0045670 |
regulation of osteoclast differentiation(GO:0045670) |
| 0.0 |
0.0 |
GO:0043653 |
mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 |
0.1 |
GO:0000338 |
protein deneddylation(GO:0000338) |
| 0.0 |
0.1 |
GO:0032431 |
activation of phospholipase A2 activity(GO:0032431) |
| 0.0 |
1.3 |
GO:0007190 |
activation of adenylate cyclase activity(GO:0007190) |
| 0.0 |
0.1 |
GO:0002032 |
desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 |
1.0 |
GO:0060349 |
bone morphogenesis(GO:0060349) |
| 0.0 |
1.4 |
GO:0048477 |
oogenesis(GO:0048477) |
| 0.0 |
0.2 |
GO:0050774 |
negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 |
0.1 |
GO:0006655 |
phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin metabolic process(GO:0032048) cardiolipin biosynthetic process(GO:0032049) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.0 |
0.2 |
GO:0006778 |
porphyrin-containing compound metabolic process(GO:0006778) tetrapyrrole metabolic process(GO:0033013) |
| 0.0 |
0.3 |
GO:0045824 |
negative regulation of innate immune response(GO:0045824) |
| 0.0 |
0.2 |
GO:0001773 |
myeloid dendritic cell activation(GO:0001773) |
| 0.0 |
0.2 |
GO:0006284 |
base-excision repair(GO:0006284) |
| 0.0 |
0.5 |
GO:0030212 |
hyaluronan metabolic process(GO:0030212) |
| 0.0 |
0.4 |
GO:0042572 |
retinol metabolic process(GO:0042572) |
| 0.0 |
0.1 |
GO:0030037 |
actin filament reorganization involved in cell cycle(GO:0030037) actin filament reorganization(GO:0090527) |
| 0.0 |
0.4 |
GO:0045070 |
positive regulation of viral genome replication(GO:0045070) |
| 0.0 |
1.6 |
GO:0051262 |
protein tetramerization(GO:0051262) |
| 0.0 |
0.4 |
GO:0007076 |
mitotic chromosome condensation(GO:0007076) |
| 0.0 |
1.0 |
GO:0051925 |
obsolete regulation of calcium ion transport via voltage-gated calcium channel activity(GO:0051925) |
| 0.0 |
0.2 |
GO:0046777 |
protein autophosphorylation(GO:0046777) |
| 0.0 |
0.1 |
GO:0030859 |
polarized epithelial cell differentiation(GO:0030859) establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 |
0.8 |
GO:0030218 |
erythrocyte differentiation(GO:0030218) |
| 0.0 |
3.1 |
GO:0006921 |
cellular component disassembly involved in execution phase of apoptosis(GO:0006921) execution phase of apoptosis(GO:0097194) |
| 0.0 |
0.1 |
GO:0006701 |
progesterone biosynthetic process(GO:0006701) |
| 0.0 |
0.1 |
GO:0030261 |
chromosome condensation(GO:0030261) |
| 0.0 |
0.1 |
GO:0033085 |
negative regulation of T cell differentiation in thymus(GO:0033085) negative regulation of thymocyte aggregation(GO:2000399) |
| 0.0 |
0.4 |
GO:0014823 |
response to activity(GO:0014823) |
| 0.0 |
0.1 |
GO:0060028 |
convergent extension involved in axis elongation(GO:0060028) negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 |
3.5 |
GO:0006333 |
chromatin assembly or disassembly(GO:0006333) |
| 0.0 |
0.7 |
GO:0006730 |
one-carbon metabolic process(GO:0006730) |
| 0.0 |
0.6 |
GO:0007214 |
gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 |
0.8 |
GO:0018022 |
peptidyl-lysine methylation(GO:0018022) |
| 0.0 |
6.1 |
GO:0000398 |
RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 |
0.5 |
GO:0050832 |
defense response to fungus(GO:0050832) |
| 0.0 |
0.0 |
GO:0060689 |
cell differentiation involved in salivary gland development(GO:0060689) |
| 0.0 |
3.0 |
GO:0015718 |
monocarboxylic acid transport(GO:0015718) |
| 0.0 |
4.0 |
GO:0006836 |
neurotransmitter transport(GO:0006836) |
| 0.0 |
0.3 |
GO:1903078 |
positive regulation of establishment of protein localization to plasma membrane(GO:0090004) positive regulation of protein localization to plasma membrane(GO:1903078) positive regulation of protein localization to cell periphery(GO:1904377) |
| 0.0 |
0.1 |
GO:0006720 |
isoprenoid metabolic process(GO:0006720) |
| 0.0 |
0.1 |
GO:0070634 |
transepithelial ammonium transport(GO:0070634) |
| 0.0 |
0.2 |
GO:0070537 |
histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 |
0.1 |
GO:0031214 |
biomineral tissue development(GO:0031214) |
| 0.0 |
4.4 |
GO:0006260 |
DNA replication(GO:0006260) |
| 0.0 |
5.2 |
GO:0006470 |
protein dephosphorylation(GO:0006470) |
| 0.0 |
0.4 |
GO:0030593 |
neutrophil chemotaxis(GO:0030593) neutrophil migration(GO:1990266) |
| 0.0 |
0.1 |
GO:0051445 |
regulation of meiotic nuclear division(GO:0040020) regulation of meiotic cell cycle(GO:0051445) |
| 0.0 |
0.3 |
GO:0051057 |
cerebral cortex tangential migration(GO:0021800) positive regulation of small GTPase mediated signal transduction(GO:0051057) |
| 0.0 |
0.3 |
GO:0044349 |
nucleotide-excision repair, DNA damage removal(GO:0000718) DNA excision(GO:0044349) |
| 0.0 |
0.2 |
GO:0043065 |
positive regulation of apoptotic process(GO:0043065) |
| 0.0 |
0.2 |
GO:0045651 |
positive regulation of macrophage differentiation(GO:0045651) positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 |
0.6 |
GO:0072332 |
intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 |
0.1 |
GO:0060136 |
embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 |
0.2 |
GO:0055015 |
ventricular cardiac muscle cell differentiation(GO:0055012) ventricular cardiac muscle cell development(GO:0055015) |
| 0.0 |
0.7 |
GO:0007605 |
sensory perception of sound(GO:0007605) |
| 0.0 |
0.6 |
GO:0019722 |
calcium-mediated signaling(GO:0019722) |
| 0.0 |
0.1 |
GO:0060992 |
response to fungicide(GO:0060992) |
| 0.0 |
0.3 |
GO:0060351 |
cartilage development involved in endochondral bone morphogenesis(GO:0060351) |
| 0.0 |
0.1 |
GO:1901503 |
ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 |
0.3 |
GO:0061351 |
neural precursor cell proliferation(GO:0061351) |
| 0.0 |
0.1 |
GO:0044872 |
lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 |
0.7 |
GO:0030520 |
intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.0 |
0.6 |
GO:0016254 |
preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 |
0.6 |
GO:0006490 |
dolichol-linked oligosaccharide biosynthetic process(GO:0006488) oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 |
0.4 |
GO:0006383 |
transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 |
1.3 |
GO:0006446 |
regulation of translational initiation(GO:0006446) |
| 0.0 |
0.2 |
GO:0030225 |
macrophage differentiation(GO:0030225) |
| 0.0 |
0.4 |
GO:0016601 |
Rac protein signal transduction(GO:0016601) |
| 0.0 |
0.2 |
GO:0048384 |
retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 |
2.8 |
GO:0010608 |
posttranscriptional regulation of gene expression(GO:0010608) |
| 0.0 |
0.1 |
GO:0060716 |
labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 |
0.1 |
GO:1903035 |
negative regulation of response to wounding(GO:1903035) |
| 0.0 |
0.1 |
GO:0021615 |
glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 |
0.9 |
GO:0009791 |
post-embryonic development(GO:0009791) |
| 0.0 |
0.2 |
GO:0015985 |
energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 |
0.1 |
GO:0048015 |
phosphatidylinositol-mediated signaling(GO:0048015) inositol lipid-mediated signaling(GO:0048017) |
| 0.0 |
0.1 |
GO:0070050 |
neuron cellular homeostasis(GO:0070050) |
| 0.0 |
0.1 |
GO:0006382 |
adenosine to inosine editing(GO:0006382) |
| 0.0 |
0.5 |
GO:0016338 |
calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 |
0.1 |
GO:0008343 |
adult feeding behavior(GO:0008343) |
| 0.0 |
0.5 |
GO:0019933 |
cAMP-mediated signaling(GO:0019933) |
| 0.0 |
0.4 |
GO:0019370 |
leukotriene biosynthetic process(GO:0019370) |
| 0.0 |
0.2 |
GO:0007020 |
microtubule nucleation(GO:0007020) |
| 0.0 |
0.8 |
GO:0031098 |
stress-activated protein kinase signaling cascade(GO:0031098) stress-activated MAPK cascade(GO:0051403) |
| 0.0 |
0.1 |
GO:0030252 |
growth hormone secretion(GO:0030252) |
| 0.0 |
0.1 |
GO:0046487 |
glyoxylate metabolic process(GO:0046487) |
| 0.0 |
0.1 |
GO:0007181 |
transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 |
0.2 |
GO:0006940 |
regulation of smooth muscle contraction(GO:0006940) |
| 0.0 |
0.1 |
GO:0018095 |
protein polyglutamylation(GO:0018095) |
| 0.0 |
0.1 |
GO:0045056 |
transcytosis(GO:0045056) |
| 0.0 |
0.1 |
GO:0070528 |
protein kinase C signaling(GO:0070528) |
| 0.0 |
0.3 |
GO:0007045 |
cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) |
| 0.0 |
0.6 |
GO:0017144 |
drug metabolic process(GO:0017144) |
| 0.0 |
1.7 |
GO:0007218 |
neuropeptide signaling pathway(GO:0007218) |
| 0.0 |
0.2 |
GO:0070389 |
chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 |
0.3 |
GO:0046323 |
glucose import(GO:0046323) |
| 0.0 |
0.0 |
GO:0072393 |
microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.0 |
1.6 |
GO:0006310 |
DNA recombination(GO:0006310) |
| 0.0 |
0.1 |
GO:2000858 |
renin-angiotensin regulation of aldosterone production(GO:0002018) regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) regulation of NAD(P)H oxidase activity(GO:0033860) positive regulation of NAD(P)H oxidase activity(GO:0033864) mineralocorticoid secretion(GO:0035931) aldosterone secretion(GO:0035932) regulation of mineralocorticoid secretion(GO:2000855) regulation of aldosterone secretion(GO:2000858) |
| 0.0 |
0.2 |
GO:0046854 |
phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 |
0.7 |
GO:0006888 |
ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 |
0.8 |
GO:0006120 |
mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 |
0.1 |
GO:0010765 |
positive regulation of sodium ion transport(GO:0010765) |
| 0.0 |
0.1 |
GO:0000060 |
protein import into nucleus, translocation(GO:0000060) |
| 0.0 |
0.1 |
GO:0045652 |
regulation of megakaryocyte differentiation(GO:0045652) regulation of hematopoietic progenitor cell differentiation(GO:1901532) |
| 0.0 |
0.1 |
GO:0002456 |
T cell mediated immunity(GO:0002456) |
| 0.0 |
0.2 |
GO:0007635 |
chemosensory behavior(GO:0007635) |
| 0.0 |
0.3 |
GO:0007398 |
ectoderm development(GO:0007398) |
| 0.0 |
0.2 |
GO:0042491 |
auditory receptor cell differentiation(GO:0042491) |
| 0.0 |
0.1 |
GO:0016998 |
cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 |
0.0 |
GO:0006471 |
protein ADP-ribosylation(GO:0006471) |
| 0.0 |
0.1 |
GO:0070293 |
renal absorption(GO:0070293) renal water absorption(GO:0070295) |
| 0.0 |
0.1 |
GO:0001649 |
osteoblast differentiation(GO:0001649) |
| 0.0 |
0.0 |
GO:0032230 |
positive regulation of synaptic transmission, GABAergic(GO:0032230) |
| 0.0 |
0.1 |
GO:0030183 |
B cell differentiation(GO:0030183) |
| 0.0 |
0.2 |
GO:0015992 |
proton transport(GO:0015992) |
| 0.0 |
0.1 |
GO:0042632 |
cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 |
0.1 |
GO:0051881 |
regulation of mitochondrial membrane potential(GO:0051881) |
| 0.0 |
0.2 |
GO:0000083 |
regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 |
0.3 |
GO:0007126 |
meiotic nuclear division(GO:0007126) |
| 0.0 |
0.0 |
GO:0071103 |
DNA conformation change(GO:0071103) |
| 0.0 |
0.3 |
GO:0007157 |
heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 |
0.2 |
GO:0030325 |
adrenal gland development(GO:0030325) |
| 0.0 |
0.1 |
GO:0060421 |
positive regulation of cardiac muscle tissue growth(GO:0055023) positive regulation of cardiac muscle tissue development(GO:0055025) positive regulation of cardiac muscle cell proliferation(GO:0060045) positive regulation of heart growth(GO:0060421) |
| 0.0 |
0.3 |
GO:0006942 |
regulation of striated muscle contraction(GO:0006942) |
| 0.0 |
0.0 |
GO:0048681 |
negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.0 |
0.2 |
GO:0070555 |
response to interleukin-1(GO:0070555) |
| 0.0 |
0.6 |
GO:0050890 |
cognition(GO:0050890) |
| 0.0 |
0.0 |
GO:0016188 |
synaptic vesicle maturation(GO:0016188) |
| 0.0 |
0.2 |
GO:0006970 |
response to osmotic stress(GO:0006970) |
| 0.0 |
0.1 |
GO:0015919 |
peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 |
0.4 |
GO:0030166 |
proteoglycan biosynthetic process(GO:0030166) |
| 0.0 |
0.3 |
GO:0006826 |
iron ion transport(GO:0006826) |
| 0.0 |
0.9 |
GO:0050851 |
antigen receptor-mediated signaling pathway(GO:0050851) |
| 0.0 |
0.1 |
GO:0016458 |
gene silencing(GO:0016458) |
| 0.0 |
2.5 |
GO:0006412 |
translation(GO:0006412) |
| 0.0 |
0.0 |
GO:0007210 |
serotonin receptor signaling pathway(GO:0007210) |
| 0.0 |
0.1 |
GO:0030901 |
midbrain development(GO:0030901) |
| 0.0 |
0.2 |
GO:0006520 |
cellular amino acid metabolic process(GO:0006520) |
| 0.0 |
0.1 |
GO:0009812 |
flavonoid metabolic process(GO:0009812) |
| 0.0 |
0.1 |
GO:0009948 |
anterior/posterior axis specification(GO:0009948) |