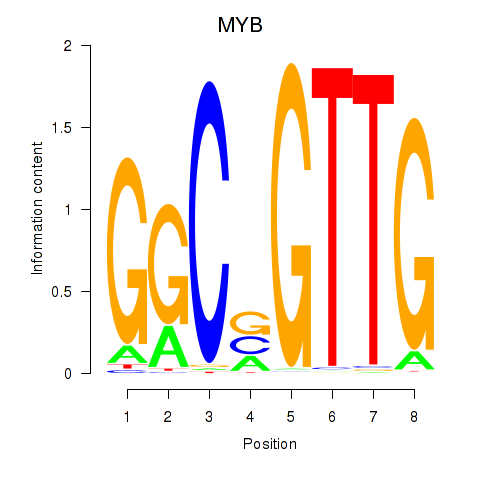
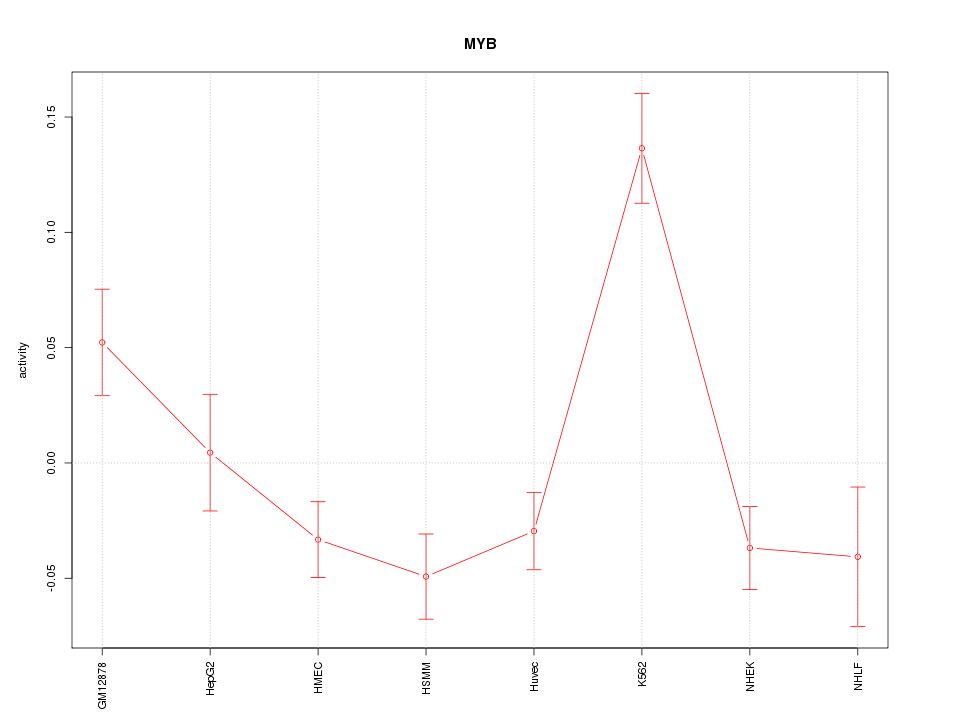
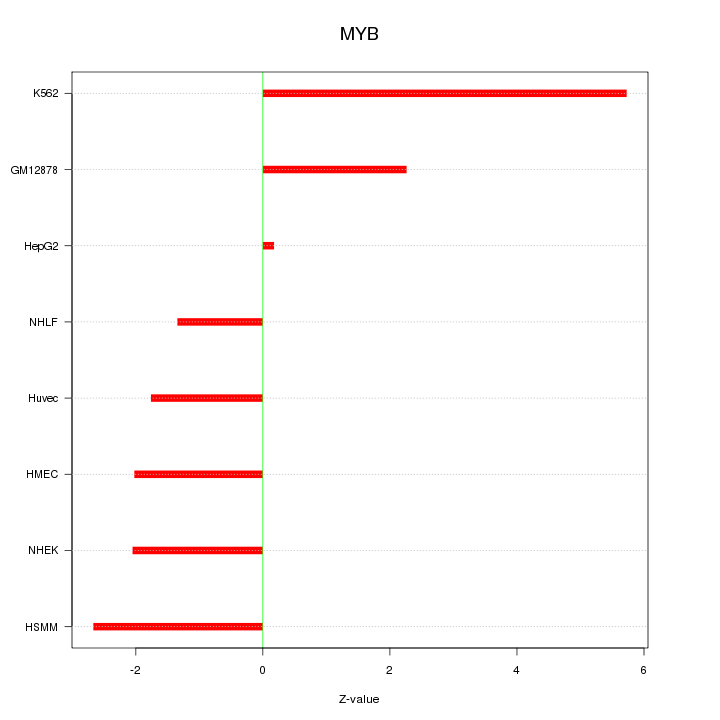
| 1.9 |
5.6 |
GO:0019695 |
choline metabolic process(GO:0019695) |
| 0.9 |
0.9 |
GO:0045048 |
protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.8 |
4.2 |
GO:0061003 |
positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.7 |
2.0 |
GO:0071539 |
protein localization to centrosome(GO:0071539) |
| 0.7 |
3.9 |
GO:0061099 |
negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.6 |
2.5 |
GO:0030259 |
lipid glycosylation(GO:0030259) |
| 0.5 |
5.2 |
GO:0016180 |
snRNA processing(GO:0016180) |
| 0.5 |
1.5 |
GO:0006530 |
asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.5 |
2.8 |
GO:0006398 |
mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.4 |
1.7 |
GO:0061032 |
visceral serous pericardium development(GO:0061032) |
| 0.4 |
2.8 |
GO:0035058 |
nonmotile primary cilium assembly(GO:0035058) |
| 0.4 |
2.9 |
GO:0006561 |
proline biosynthetic process(GO:0006561) |
| 0.3 |
2.0 |
GO:0051299 |
centrosome separation(GO:0051299) |
| 0.3 |
2.0 |
GO:0043316 |
natural killer cell activation involved in immune response(GO:0002323) cytotoxic T cell degranulation(GO:0043316) natural killer cell degranulation(GO:0043320) |
| 0.3 |
0.7 |
GO:0010826 |
negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.3 |
0.6 |
GO:0032071 |
regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.3 |
0.9 |
GO:0007141 |
male meiosis I(GO:0007141) |
| 0.3 |
0.8 |
GO:0031077 |
post-embryonic camera-type eye development(GO:0031077) Spemann organizer formation(GO:0060061) |
| 0.3 |
2.5 |
GO:0007063 |
regulation of sister chromatid cohesion(GO:0007063) |
| 0.3 |
0.8 |
GO:0000338 |
protein deneddylation(GO:0000338) |
| 0.3 |
1.1 |
GO:0051013 |
microtubule severing(GO:0051013) |
| 0.3 |
1.1 |
GO:0000463 |
maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) maturation of LSU-rRNA(GO:0000470) |
| 0.3 |
3.8 |
GO:0070059 |
intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0070059) |
| 0.3 |
1.0 |
GO:0048025 |
negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.3 |
2.1 |
GO:0046831 |
regulation of RNA export from nucleus(GO:0046831) |
| 0.3 |
2.6 |
GO:0046784 |
viral mRNA export from host cell nucleus(GO:0046784) |
| 0.2 |
0.7 |
GO:0002326 |
B cell lineage commitment(GO:0002326) |
| 0.2 |
5.0 |
GO:0032781 |
positive regulation of ATPase activity(GO:0032781) |
| 0.2 |
1.4 |
GO:0046599 |
regulation of centriole replication(GO:0046599) |
| 0.2 |
0.7 |
GO:0072677 |
eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.2 |
1.7 |
GO:0051096 |
meiotic mismatch repair(GO:0000710) positive regulation of helicase activity(GO:0051096) |
| 0.2 |
1.4 |
GO:0070934 |
CRD-mediated mRNA stabilization(GO:0070934) |
| 0.2 |
0.8 |
GO:0040016 |
embryonic cleavage(GO:0040016) |
| 0.2 |
1.7 |
GO:1900087 |
traversing start control point of mitotic cell cycle(GO:0007089) positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) positive regulation of cell cycle G1/S phase transition(GO:1902808) |
| 0.2 |
0.2 |
GO:0045116 |
protein neddylation(GO:0045116) |
| 0.2 |
1.1 |
GO:0019985 |
translesion synthesis(GO:0019985) |
| 0.2 |
0.9 |
GO:0070935 |
3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.2 |
3.7 |
GO:0043968 |
histone H2A acetylation(GO:0043968) |
| 0.2 |
1.0 |
GO:0000089 |
mitotic metaphase(GO:0000089) |
| 0.2 |
0.9 |
GO:0031936 |
negative regulation of chromatin silencing(GO:0031936) |
| 0.2 |
0.7 |
GO:0044321 |
cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.2 |
1.0 |
GO:0006422 |
aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.2 |
2.2 |
GO:0007076 |
mitotic chromosome condensation(GO:0007076) |
| 0.2 |
1.4 |
GO:0048387 |
negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.2 |
0.8 |
GO:0032933 |
SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.2 |
0.6 |
GO:0001807 |
type IV hypersensitivity(GO:0001806) regulation of type IV hypersensitivity(GO:0001807) |
| 0.2 |
1.7 |
GO:0006776 |
vitamin A metabolic process(GO:0006776) |
| 0.2 |
0.5 |
GO:0003289 |
septum primum development(GO:0003284) atrial septum primum morphogenesis(GO:0003289) |
| 0.2 |
0.5 |
GO:0060316 |
positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.2 |
5.0 |
GO:0000387 |
spliceosomal snRNP assembly(GO:0000387) |
| 0.2 |
0.6 |
GO:0035519 |
protein K29-linked ubiquitination(GO:0035519) |
| 0.1 |
0.6 |
GO:0043923 |
positive regulation by host of viral transcription(GO:0043923) |
| 0.1 |
2.1 |
GO:0006449 |
regulation of translational termination(GO:0006449) |
| 0.1 |
0.4 |
GO:0014889 |
muscle atrophy(GO:0014889) |
| 0.1 |
0.4 |
GO:0070407 |
oxidation-dependent protein catabolic process(GO:0070407) |
| 0.1 |
1.9 |
GO:0020027 |
hemoglobin metabolic process(GO:0020027) |
| 0.1 |
0.8 |
GO:0000729 |
DNA double-strand break processing(GO:0000729) |
| 0.1 |
0.4 |
GO:0014022 |
neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.1 |
0.4 |
GO:0006106 |
fumarate metabolic process(GO:0006106) |
| 0.1 |
0.4 |
GO:0035408 |
histone H3-T6 phosphorylation(GO:0035408) |
| 0.1 |
0.8 |
GO:0009082 |
branched-chain amino acid biosynthetic process(GO:0009082) |
| 0.1 |
0.8 |
GO:0048617 |
embryonic foregut morphogenesis(GO:0048617) |
| 0.1 |
0.4 |
GO:0048262 |
determination of dorsal/ventral asymmetry(GO:0048262) determination of dorsal identity(GO:0048263) |
| 0.1 |
0.5 |
GO:0001840 |
neural plate morphogenesis(GO:0001839) neural plate development(GO:0001840) |
| 0.1 |
0.6 |
GO:0060136 |
embryonic process involved in female pregnancy(GO:0060136) |
| 0.1 |
0.7 |
GO:0046898 |
response to cycloheximide(GO:0046898) |
| 0.1 |
0.5 |
GO:0051958 |
methotrexate transport(GO:0051958) |
| 0.1 |
1.1 |
GO:0015937 |
coenzyme A biosynthetic process(GO:0015937) |
| 0.1 |
1.0 |
GO:0010507 |
negative regulation of autophagy(GO:0010507) |
| 0.1 |
1.3 |
GO:0051322 |
anaphase(GO:0051322) |
| 0.1 |
0.5 |
GO:0032042 |
mitochondrial DNA replication(GO:0006264) mitochondrial DNA metabolic process(GO:0032042) |
| 0.1 |
0.8 |
GO:0016998 |
cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 |
1.1 |
GO:0045824 |
negative regulation of innate immune response(GO:0045824) |
| 0.1 |
0.3 |
GO:0033211 |
adiponectin-activated signaling pathway(GO:0033211) |
| 0.1 |
0.3 |
GO:0036037 |
CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 |
0.5 |
GO:0060164 |
regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 |
1.4 |
GO:0007020 |
microtubule nucleation(GO:0007020) |
| 0.1 |
0.9 |
GO:0071108 |
protein K48-linked deubiquitination(GO:0071108) |
| 0.1 |
1.7 |
GO:0045742 |
positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) positive regulation of ERBB signaling pathway(GO:1901186) |
| 0.1 |
0.4 |
GO:0061303 |
cornea development in camera-type eye(GO:0061303) |
| 0.1 |
0.4 |
GO:0048845 |
venous blood vessel morphogenesis(GO:0048845) |
| 0.1 |
1.8 |
GO:0000038 |
very long-chain fatty acid metabolic process(GO:0000038) |
| 0.1 |
0.8 |
GO:0048712 |
negative regulation of astrocyte differentiation(GO:0048712) |
| 0.1 |
0.3 |
GO:0021527 |
spinal cord association neuron differentiation(GO:0021527) neuron fate determination(GO:0048664) |
| 0.1 |
0.4 |
GO:0032696 |
negative regulation of interleukin-13 production(GO:0032696) regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) regulation of receptor binding(GO:1900120) negative regulation of receptor binding(GO:1900121) |
| 0.1 |
1.0 |
GO:0060762 |
regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.1 |
0.2 |
GO:0048680 |
astrocyte activation(GO:0048143) positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.1 |
1.1 |
GO:0043097 |
pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.1 |
0.2 |
GO:0002295 |
T-helper cell lineage commitment(GO:0002295) alpha-beta T cell lineage commitment(GO:0002363) CD4-positive or CD8-positive, alpha-beta T cell lineage commitment(GO:0043369) CD4-positive, alpha-beta T cell lineage commitment(GO:0043373) memory T cell differentiation(GO:0043379) regulation of memory T cell differentiation(GO:0043380) positive regulation of memory T cell differentiation(GO:0043382) T-helper 17 type immune response(GO:0072538) T-helper 17 cell differentiation(GO:0072539) T-helper 17 cell lineage commitment(GO:0072540) regulation of T-helper 17 type immune response(GO:2000316) positive regulation of T-helper 17 type immune response(GO:2000318) regulation of T-helper 17 cell differentiation(GO:2000319) positive regulation of T-helper 17 cell differentiation(GO:2000321) regulation of T-helper 17 cell lineage commitment(GO:2000328) positive regulation of T-helper 17 cell lineage commitment(GO:2000330) |
| 0.1 |
0.9 |
GO:0006027 |
glycosaminoglycan catabolic process(GO:0006027) |
| 0.1 |
0.7 |
GO:0015074 |
DNA integration(GO:0015074) |
| 0.1 |
0.3 |
GO:0042816 |
pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.1 |
1.0 |
GO:0007096 |
regulation of exit from mitosis(GO:0007096) |
| 0.1 |
0.1 |
GO:0042701 |
progesterone secretion(GO:0042701) |
| 0.1 |
0.7 |
GO:0010452 |
histone H3-K36 methylation(GO:0010452) |
| 0.1 |
2.3 |
GO:0042744 |
hydrogen peroxide catabolic process(GO:0042744) |
| 0.1 |
0.8 |
GO:0000076 |
DNA replication checkpoint(GO:0000076) |
| 0.1 |
0.4 |
GO:0002051 |
osteoblast fate commitment(GO:0002051) |
| 0.1 |
0.4 |
GO:0060707 |
trophoblast giant cell differentiation(GO:0060707) |
| 0.1 |
0.5 |
GO:0045010 |
actin nucleation(GO:0045010) |
| 0.1 |
0.5 |
GO:0016075 |
rRNA catabolic process(GO:0016075) |
| 0.1 |
0.4 |
GO:0042769 |
DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 |
0.7 |
GO:0090090 |
negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.1 |
0.6 |
GO:0032148 |
activation of protein kinase B activity(GO:0032148) |
| 0.1 |
0.5 |
GO:0015853 |
adenine transport(GO:0015853) |
| 0.1 |
0.7 |
GO:0048013 |
ephrin receptor signaling pathway(GO:0048013) |
| 0.1 |
1.0 |
GO:0050930 |
induction of positive chemotaxis(GO:0050930) |
| 0.1 |
2.2 |
GO:0016925 |
protein sumoylation(GO:0016925) |
| 0.1 |
2.8 |
GO:0006369 |
termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 |
0.7 |
GO:0008634 |
obsolete negative regulation of survival gene product expression(GO:0008634) |
| 0.1 |
0.8 |
GO:0050684 |
regulation of mRNA processing(GO:0050684) regulation of mRNA metabolic process(GO:1903311) |
| 0.1 |
1.7 |
GO:0015949 |
nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.1 |
0.6 |
GO:0006268 |
DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 |
0.4 |
GO:0070498 |
response to peptidoglycan(GO:0032494) interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.1 |
0.8 |
GO:0070373 |
negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.1 |
0.2 |
GO:0015917 |
aminophospholipid transport(GO:0015917) |
| 0.1 |
1.5 |
GO:0043278 |
response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.1 |
1.0 |
GO:0000380 |
alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.1 |
0.2 |
GO:0061084 |
regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) negative regulation of protein folding(GO:1903333) |
| 0.1 |
1.5 |
GO:0042531 |
positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.1 |
2.2 |
GO:1901799 |
negative regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032435) negative regulation of proteasomal protein catabolic process(GO:1901799) |
| 0.1 |
0.8 |
GO:0070534 |
protein K63-linked ubiquitination(GO:0070534) |
| 0.1 |
0.6 |
GO:0001539 |
cilium or flagellum-dependent cell motility(GO:0001539) |
| 0.0 |
1.4 |
GO:0017144 |
drug metabolic process(GO:0017144) |
| 0.0 |
0.1 |
GO:0045759 |
negative regulation of action potential(GO:0045759) |
| 0.0 |
0.4 |
GO:0043545 |
Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 |
0.9 |
GO:0048246 |
macrophage chemotaxis(GO:0048246) |
| 0.0 |
0.3 |
GO:0007064 |
mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 |
0.6 |
GO:0040023 |
establishment of nucleus localization(GO:0040023) |
| 0.0 |
1.3 |
GO:0007052 |
mitotic spindle organization(GO:0007052) |
| 0.0 |
0.3 |
GO:0045008 |
depyrimidination(GO:0045008) |
| 0.0 |
0.9 |
GO:0032092 |
positive regulation of protein binding(GO:0032092) |
| 0.0 |
0.7 |
GO:0051703 |
social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 |
1.3 |
GO:0071804 |
cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 |
0.2 |
GO:0005981 |
regulation of glycogen catabolic process(GO:0005981) |
| 0.0 |
6.5 |
GO:0000398 |
RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 |
0.4 |
GO:0030968 |
endoplasmic reticulum unfolded protein response(GO:0030968) cellular response to unfolded protein(GO:0034620) cellular response to topologically incorrect protein(GO:0035967) |
| 0.0 |
1.1 |
GO:0006362 |
transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 |
0.2 |
GO:0046655 |
folic acid metabolic process(GO:0046655) |
| 0.0 |
0.3 |
GO:0035235 |
ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 |
0.1 |
GO:0042727 |
flavin-containing compound biosynthetic process(GO:0042727) |
| 0.0 |
0.1 |
GO:0051967 |
negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 |
0.6 |
GO:0006098 |
pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.0 |
0.2 |
GO:0018026 |
peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 |
0.5 |
GO:0002021 |
response to dietary excess(GO:0002021) |
| 0.0 |
1.2 |
GO:0006730 |
one-carbon metabolic process(GO:0006730) |
| 0.0 |
0.4 |
GO:0051319 |
mitotic G2 phase(GO:0000085) G2 phase(GO:0051319) |
| 0.0 |
0.7 |
GO:0090630 |
activation of GTPase activity(GO:0090630) |
| 0.0 |
0.2 |
GO:0060346 |
bone trabecula formation(GO:0060346) bone trabecula morphogenesis(GO:0061430) |
| 0.0 |
0.4 |
GO:0031958 |
corticosteroid receptor signaling pathway(GO:0031958) glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.0 |
0.4 |
GO:0030388 |
fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 |
0.2 |
GO:0051298 |
centrosome duplication(GO:0051298) |
| 0.0 |
0.8 |
GO:0030514 |
negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 |
0.2 |
GO:0035162 |
embryonic hemopoiesis(GO:0035162) |
| 0.0 |
1.1 |
GO:0006284 |
base-excision repair(GO:0006284) |
| 0.0 |
0.2 |
GO:0042474 |
middle ear morphogenesis(GO:0042474) |
| 0.0 |
0.5 |
GO:0045749 |
obsolete negative regulation of S phase of mitotic cell cycle(GO:0045749) |
| 0.0 |
0.4 |
GO:0030336 |
negative regulation of cell migration(GO:0030336) negative regulation of cell motility(GO:2000146) |
| 0.0 |
0.2 |
GO:0006893 |
Golgi to plasma membrane transport(GO:0006893) vesicle-mediated transport to the plasma membrane(GO:0098876) |
| 0.0 |
0.3 |
GO:0032364 |
oxygen homeostasis(GO:0032364) |
| 0.0 |
1.7 |
GO:0070588 |
calcium ion transmembrane transport(GO:0070588) |
| 0.0 |
1.9 |
GO:0030218 |
erythrocyte differentiation(GO:0030218) |
| 0.0 |
0.5 |
GO:0000291 |
nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 |
1.2 |
GO:0001837 |
epithelial to mesenchymal transition(GO:0001837) |
| 0.0 |
0.5 |
GO:0000060 |
protein import into nucleus, translocation(GO:0000060) |
| 0.0 |
0.2 |
GO:0060716 |
labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 |
0.2 |
GO:0045793 |
positive regulation of cell size(GO:0045793) |
| 0.0 |
0.3 |
GO:0030041 |
actin filament polymerization(GO:0030041) |
| 0.0 |
0.1 |
GO:0007210 |
serotonin receptor signaling pathway(GO:0007210) |
| 0.0 |
0.2 |
GO:0000239 |
pachytene(GO:0000239) |
| 0.0 |
0.4 |
GO:0018298 |
protein-chromophore linkage(GO:0018298) |
| 0.0 |
1.9 |
GO:0006413 |
translational initiation(GO:0006413) |
| 0.0 |
0.1 |
GO:0006782 |
protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.0 |
0.6 |
GO:0042147 |
retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 |
1.2 |
GO:0044839 |
G2/M transition of mitotic cell cycle(GO:0000086) cell cycle G2/M phase transition(GO:0044839) |
| 0.0 |
0.7 |
GO:0070830 |
bicellular tight junction assembly(GO:0070830) |
| 0.0 |
0.6 |
GO:1901343 |
negative regulation of angiogenesis(GO:0016525) negative regulation of vasculature development(GO:1901343) negative regulation of blood vessel morphogenesis(GO:2000181) |
| 0.0 |
1.2 |
GO:0050680 |
negative regulation of epithelial cell proliferation(GO:0050680) |
| 0.0 |
0.2 |
GO:0048536 |
spleen development(GO:0048536) |
| 0.0 |
0.5 |
GO:0031623 |
receptor internalization(GO:0031623) |
| 0.0 |
0.2 |
GO:0001947 |
heart looping(GO:0001947) embryonic heart tube morphogenesis(GO:0003143) determination of heart left/right asymmetry(GO:0061371) |
| 0.0 |
0.3 |
GO:0006406 |
mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 |
0.2 |
GO:0034379 |
very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.0 |
0.2 |
GO:0046689 |
response to mercury ion(GO:0046689) |
| 0.0 |
0.6 |
GO:0043966 |
histone H3 acetylation(GO:0043966) |
| 0.0 |
0.2 |
GO:0046718 |
viral entry into host cell(GO:0046718) |
| 0.0 |
0.2 |
GO:0006465 |
signal peptide processing(GO:0006465) |
| 0.0 |
0.1 |
GO:0048537 |
mucosal-associated lymphoid tissue development(GO:0048537) |
| 0.0 |
0.7 |
GO:0006888 |
ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 |
0.3 |
GO:0006611 |
protein export from nucleus(GO:0006611) |
| 0.0 |
0.1 |
GO:0009649 |
entrainment of circadian clock(GO:0009649) |
| 0.0 |
0.1 |
GO:0072164 |
ureteric bud development(GO:0001657) mesonephric epithelium development(GO:0072163) mesonephric tubule development(GO:0072164) |
| 0.0 |
0.2 |
GO:0033523 |
histone monoubiquitination(GO:0010390) histone H2B ubiquitination(GO:0033523) |
| 0.0 |
0.1 |
GO:0006474 |
N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 |
0.1 |
GO:0042976 |
activation of Janus kinase activity(GO:0042976) activation of JAK2 kinase activity(GO:0042977) |
| 0.0 |
0.2 |
GO:1902603 |
carnitine shuttle(GO:0006853) carnitine transmembrane transport(GO:1902603) |
| 0.0 |
1.6 |
GO:0006897 |
endocytosis(GO:0006897) |
| 0.0 |
0.5 |
GO:0009060 |
aerobic respiration(GO:0009060) |
| 0.0 |
0.1 |
GO:0042354 |
fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 |
0.9 |
GO:0008360 |
regulation of cell shape(GO:0008360) |
| 0.0 |
0.2 |
GO:0006044 |
N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 |
1.0 |
GO:0030832 |
regulation of actin filament length(GO:0030832) |
| 0.0 |
0.0 |
GO:0060710 |
chorio-allantoic fusion(GO:0060710) |
| 0.0 |
0.4 |
GO:0048593 |
camera-type eye morphogenesis(GO:0048593) |
| 0.0 |
0.1 |
GO:0048484 |
enteric nervous system development(GO:0048484) |
| 0.0 |
0.7 |
GO:0045666 |
positive regulation of neuron differentiation(GO:0045666) |
| 0.0 |
0.8 |
GO:0006338 |
chromatin remodeling(GO:0006338) |
| 0.0 |
1.4 |
GO:0008654 |
phospholipid biosynthetic process(GO:0008654) |
| 0.0 |
0.1 |
GO:0046349 |
UDP-N-acetylglucosamine biosynthetic process(GO:0006048) amino sugar biosynthetic process(GO:0046349) |
| 0.0 |
0.0 |
GO:0001550 |
ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |