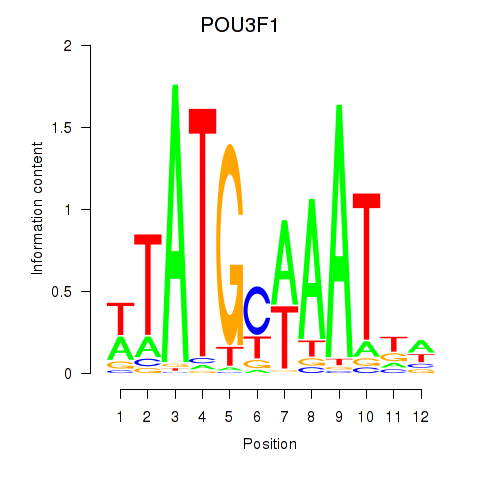
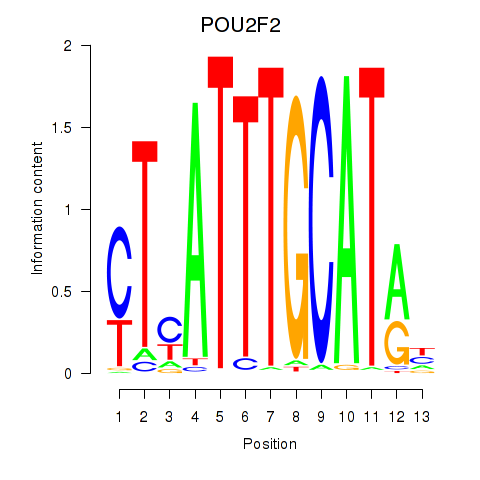
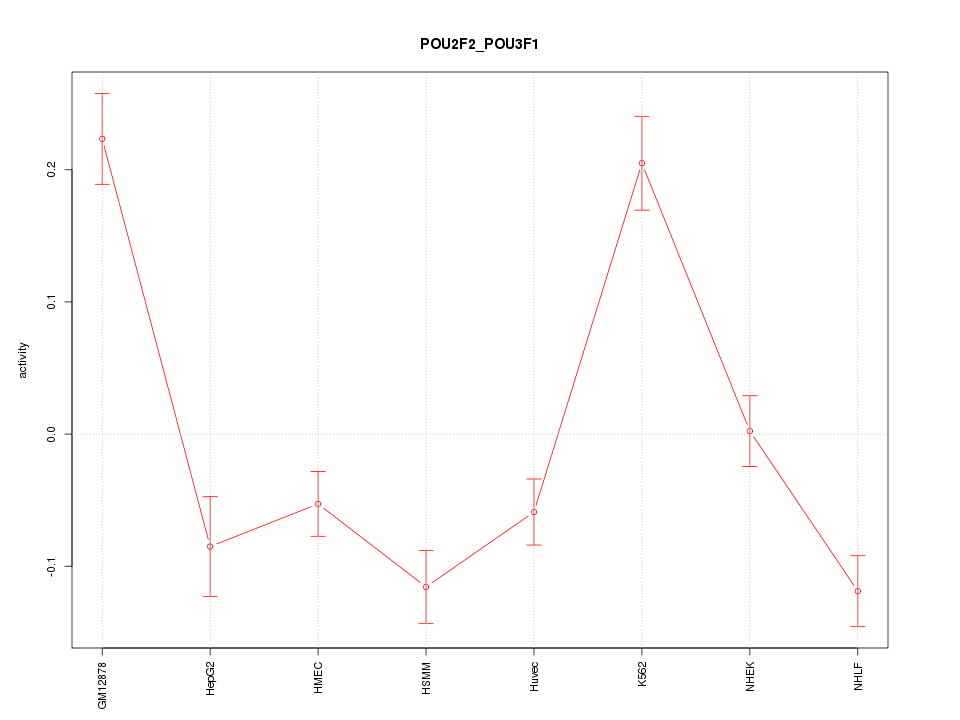
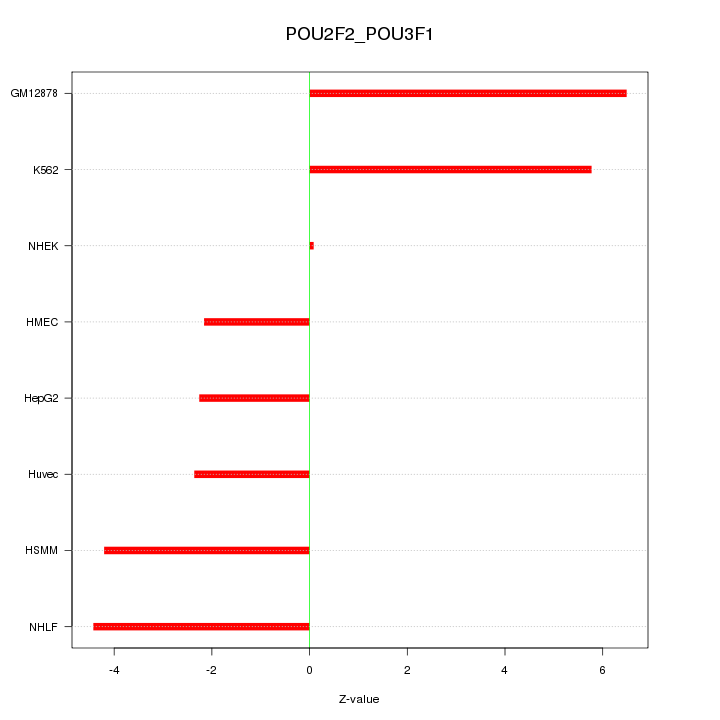
| 16.8 |
50.4 |
GO:0035408 |
histone H3-T6 phosphorylation(GO:0035408) |
| 6.5 |
19.4 |
GO:0070445 |
Fc receptor mediated stimulatory signaling pathway(GO:0002431) regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 1.8 |
9.0 |
GO:0010508 |
positive regulation of autophagy(GO:0010508) |
| 1.0 |
7.0 |
GO:0042989 |
sequestering of actin monomers(GO:0042989) |
| 0.8 |
4.1 |
GO:0034371 |
positive regulation of sequestering of triglyceride(GO:0010890) chylomicron remodeling(GO:0034371) |
| 0.8 |
75.8 |
GO:0006334 |
nucleosome assembly(GO:0006334) |
| 0.6 |
1.7 |
GO:0008050 |
female courtship behavior(GO:0008050) |
| 0.5 |
3.1 |
GO:0071699 |
olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.4 |
3.1 |
GO:0031657 |
regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031657) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031659) |
| 0.4 |
4.6 |
GO:0006776 |
vitamin A metabolic process(GO:0006776) |
| 0.4 |
1.6 |
GO:0031936 |
negative regulation of chromatin silencing(GO:0031936) |
| 0.4 |
2.7 |
GO:0021830 |
interneuron migration from the subpallium to the cortex(GO:0021830) cerebral cortex GABAergic interneuron migration(GO:0021853) cerebral cortex GABAergic interneuron development(GO:0021894) interneuron migration(GO:1904936) |
| 0.4 |
1.1 |
GO:0046984 |
regulation of hemoglobin biosynthetic process(GO:0046984) |
| 0.3 |
1.2 |
GO:0033262 |
regulation of nuclear cell cycle DNA replication(GO:0033262) replication fork protection(GO:0048478) |
| 0.3 |
1.7 |
GO:0006422 |
aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.2 |
1.5 |
GO:0006658 |
phosphatidylserine metabolic process(GO:0006658) |
| 0.2 |
1.6 |
GO:0045627 |
regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.2 |
0.7 |
GO:0019858 |
cytosine metabolic process(GO:0019858) |
| 0.2 |
0.9 |
GO:0060261 |
positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.2 |
8.3 |
GO:0065004 |
protein-DNA complex assembly(GO:0065004) |
| 0.2 |
1.2 |
GO:0030185 |
nitric oxide transport(GO:0030185) |
| 0.2 |
2.5 |
GO:0043330 |
response to exogenous dsRNA(GO:0043330) type I interferon biosynthetic process(GO:0045351) |
| 0.1 |
0.7 |
GO:0007288 |
sperm axoneme assembly(GO:0007288) |
| 0.1 |
1.3 |
GO:0016180 |
snRNA processing(GO:0016180) |
| 0.1 |
0.4 |
GO:0021913 |
regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.1 |
0.9 |
GO:0005984 |
disaccharide metabolic process(GO:0005984) |
| 0.1 |
0.5 |
GO:0034310 |
primary alcohol catabolic process(GO:0034310) |
| 0.1 |
0.6 |
GO:0007100 |
mitotic centrosome separation(GO:0007100) |
| 0.1 |
0.4 |
GO:2000001 |
regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 |
16.9 |
GO:0006959 |
humoral immune response(GO:0006959) |
| 0.1 |
1.0 |
GO:0006983 |
ER overload response(GO:0006983) |
| 0.1 |
0.4 |
GO:0031580 |
membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 |
0.5 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 |
0.6 |
GO:0060670 |
branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.1 |
0.5 |
GO:0045767 |
obsolete regulation of anti-apoptosis(GO:0045767) |
| 0.1 |
1.3 |
GO:0060736 |
prostate gland growth(GO:0060736) |
| 0.1 |
0.8 |
GO:0016045 |
detection of bacterium(GO:0016045) |
| 0.1 |
0.4 |
GO:0072369 |
positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.1 |
0.2 |
GO:0072201 |
negative regulation of mesenchymal cell proliferation(GO:0072201) |
| 0.1 |
0.4 |
GO:0009642 |
response to light intensity(GO:0009642) |
| 0.1 |
0.8 |
GO:0070327 |
thyroid hormone transport(GO:0070327) |
| 0.1 |
1.1 |
GO:0030539 |
male genitalia development(GO:0030539) |
| 0.1 |
0.9 |
GO:0009223 |
pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.1 |
0.3 |
GO:0045872 |
regulation of rhodopsin gene expression(GO:0007468) positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.1 |
0.3 |
GO:0070933 |
histone H3 deacetylation(GO:0070932) histone H4 deacetylation(GO:0070933) |
| 0.1 |
0.5 |
GO:0046835 |
carbohydrate phosphorylation(GO:0046835) |
| 0.1 |
1.0 |
GO:0032438 |
melanosome organization(GO:0032438) pigment granule organization(GO:0048753) |
| 0.1 |
6.1 |
GO:0071357 |
response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.1 |
0.3 |
GO:0032736 |
positive regulation of interleukin-13 production(GO:0032736) |
| 0.1 |
1.3 |
GO:0090399 |
replicative senescence(GO:0090399) |
| 0.1 |
0.3 |
GO:0040016 |
embryonic cleavage(GO:0040016) |
| 0.1 |
0.2 |
GO:0043276 |
anoikis(GO:0043276) |
| 0.1 |
0.3 |
GO:0032878 |
regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.1 |
1.0 |
GO:0007020 |
microtubule nucleation(GO:0007020) |
| 0.1 |
1.1 |
GO:0000380 |
alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.1 |
1.3 |
GO:0000291 |
nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.1 |
0.3 |
GO:0006003 |
fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 |
1.4 |
GO:0018208 |
peptidyl-proline modification(GO:0018208) |
| 0.0 |
0.1 |
GO:0048865 |
stem cell fate commitment(GO:0048865) stem cell fate determination(GO:0048867) |
| 0.0 |
0.5 |
GO:0048488 |
synaptic vesicle recycling(GO:0036465) synaptic vesicle endocytosis(GO:0048488) clathrin-mediated endocytosis(GO:0072583) |
| 0.0 |
0.6 |
GO:0046827 |
positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 |
0.2 |
GO:0070050 |
neuron cellular homeostasis(GO:0070050) |
| 0.0 |
0.2 |
GO:0006556 |
S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 |
0.6 |
GO:0002076 |
osteoblast development(GO:0002076) |
| 0.0 |
0.1 |
GO:0045591 |
positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.0 |
0.8 |
GO:0060113 |
inner ear receptor cell differentiation(GO:0060113) |
| 0.0 |
1.1 |
GO:0007214 |
gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 |
0.2 |
GO:0015889 |
cobalamin transport(GO:0015889) |
| 0.0 |
0.7 |
GO:0071804 |
cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 |
1.8 |
GO:0016579 |
protein deubiquitination(GO:0016579) |
| 0.0 |
0.8 |
GO:0030520 |
intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.0 |
0.4 |
GO:0008635 |
activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 |
0.5 |
GO:0030097 |
hemopoiesis(GO:0030097) |
| 0.0 |
0.1 |
GO:0010842 |
retina layer formation(GO:0010842) |
| 0.0 |
0.4 |
GO:0060711 |
labyrinthine layer development(GO:0060711) |
| 0.0 |
0.1 |
GO:0007352 |
zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 |
0.4 |
GO:0021846 |
cell proliferation in forebrain(GO:0021846) |
| 0.0 |
1.8 |
GO:0006094 |
gluconeogenesis(GO:0006094) |
| 0.0 |
0.8 |
GO:0043484 |
regulation of RNA splicing(GO:0043484) |
| 0.0 |
0.1 |
GO:0007196 |
adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 |
0.3 |
GO:0042273 |
ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 |
1.7 |
GO:0046777 |
protein autophosphorylation(GO:0046777) |
| 0.0 |
0.1 |
GO:0000460 |
maturation of 5.8S rRNA(GO:0000460) |
| 0.0 |
0.5 |
GO:0006665 |
sphingolipid metabolic process(GO:0006665) |
| 0.0 |
1.2 |
GO:0051591 |
response to cAMP(GO:0051591) |
| 0.0 |
2.5 |
GO:0051028 |
mRNA transport(GO:0051028) |
| 0.0 |
0.2 |
GO:0071300 |
cellular response to retinoic acid(GO:0071300) |
| 0.0 |
0.1 |
GO:0015937 |
coenzyme A biosynthetic process(GO:0015937) |
| 0.0 |
0.5 |
GO:0035272 |
exocrine system development(GO:0035272) |