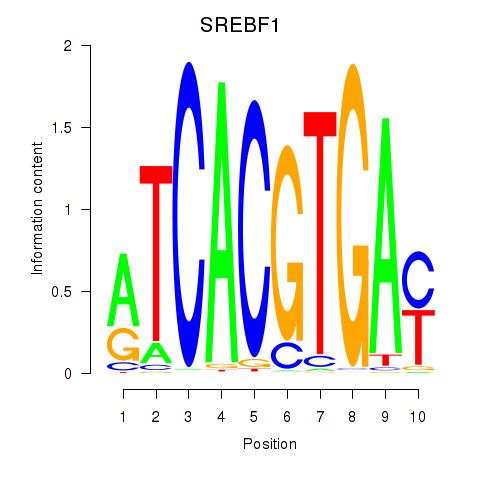
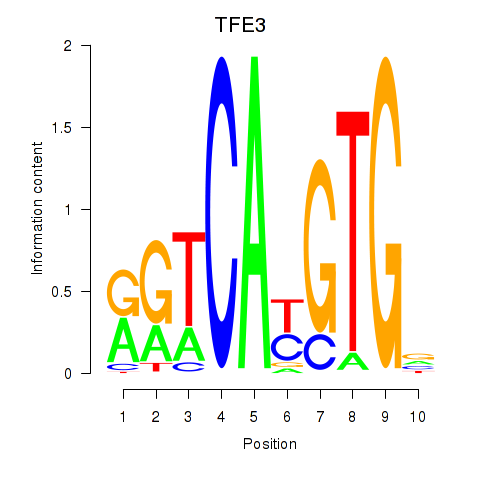
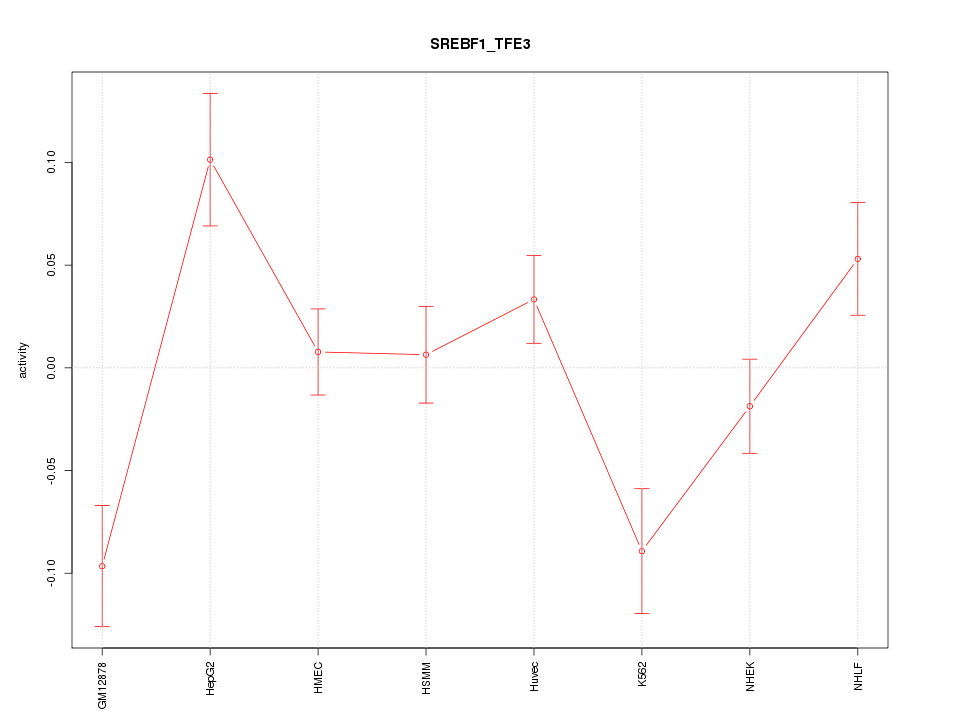
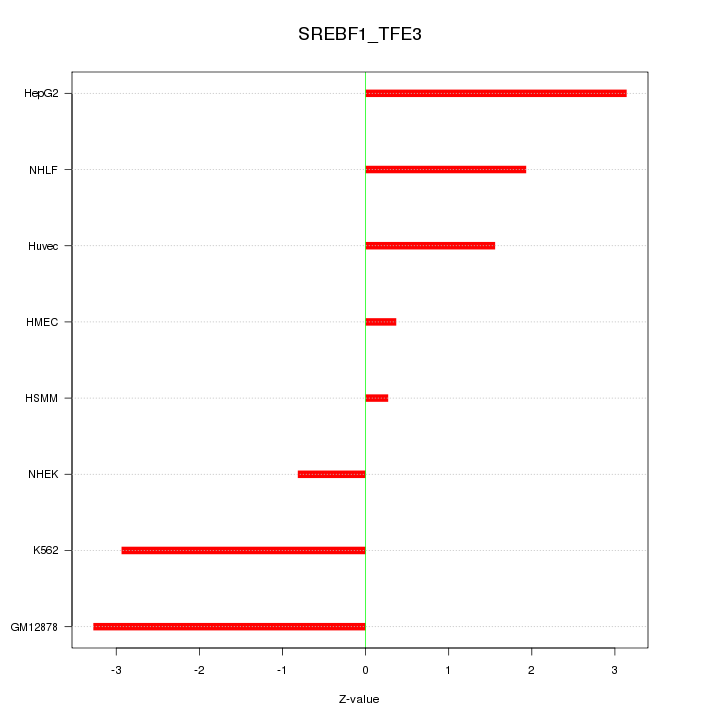
| 3.2 |
9.6 |
GO:0003011 |
diaphragm contraction(GO:0002086) involuntary skeletal muscle contraction(GO:0003011) |
| 2.1 |
4.2 |
GO:0010046 |
response to mycotoxin(GO:0010046) |
| 1.7 |
5.1 |
GO:0043371 |
negative regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043371) negative regulation of T-helper cell differentiation(GO:0045623) negative regulation of CD4-positive, alpha-beta T cell activation(GO:2000515) |
| 1.6 |
4.9 |
GO:0014022 |
neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 1.6 |
4.8 |
GO:0010248 |
establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 1.3 |
5.1 |
GO:0009609 |
response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 1.2 |
4.9 |
GO:0033986 |
response to methanol(GO:0033986) response to chromate(GO:0046687) |
| 1.1 |
10.1 |
GO:0006622 |
protein targeting to lysosome(GO:0006622) protein targeting to vacuole(GO:0006623) protein localization to lysosome(GO:0061462) protein localization to vacuole(GO:0072665) establishment of protein localization to vacuole(GO:0072666) |
| 0.9 |
2.6 |
GO:2000910 |
regulation of cholesterol import(GO:0060620) negative regulation of cholesterol import(GO:0060621) regulation of sterol import(GO:2000909) negative regulation of sterol import(GO:2000910) |
| 0.8 |
8.4 |
GO:0010739 |
positive regulation of protein kinase A signaling(GO:0010739) |
| 0.7 |
4.5 |
GO:0046882 |
negative regulation of gonadotropin secretion(GO:0032277) negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.7 |
2.1 |
GO:0002887 |
negative regulation of myeloid leukocyte mediated immunity(GO:0002887) heme oxidation(GO:0006788) smooth muscle hyperplasia(GO:0014806) muscle hyperplasia(GO:0014900) negative regulation of leukocyte degranulation(GO:0043301) |
| 0.6 |
2.3 |
GO:0001575 |
globoside metabolic process(GO:0001575) |
| 0.6 |
1.7 |
GO:0070092 |
regulation of glucagon secretion(GO:0070092) |
| 0.5 |
1.6 |
GO:1990170 |
detoxification of cadmium ion(GO:0071585) stress response to cadmium ion(GO:1990170) |
| 0.5 |
0.5 |
GO:0055098 |
response to low-density lipoprotein particle(GO:0055098) |
| 0.5 |
1.5 |
GO:0019482 |
beta-alanine metabolic process(GO:0019482) |
| 0.5 |
4.4 |
GO:0006685 |
sphingomyelin catabolic process(GO:0006685) |
| 0.5 |
1.5 |
GO:0060291 |
long-term synaptic potentiation(GO:0060291) |
| 0.5 |
1.4 |
GO:0006982 |
response to lipid hydroperoxide(GO:0006982) |
| 0.5 |
2.8 |
GO:0032007 |
negative regulation of TOR signaling(GO:0032007) |
| 0.5 |
1.4 |
GO:0060574 |
intestinal epithelial cell maturation(GO:0060574) intestinal epithelial cell development(GO:0060576) |
| 0.4 |
6.1 |
GO:0030212 |
hyaluronan metabolic process(GO:0030212) |
| 0.4 |
1.3 |
GO:0048389 |
intermediate mesoderm development(GO:0048389) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) bud dilation involved in lung branching(GO:0060503) branch elongation involved in ureteric bud branching(GO:0060681) regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) pulmonary artery morphogenesis(GO:0061156) cell proliferation involved in mesonephros development(GO:0061209) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) negative regulation of mesonephros development(GO:0061218) pattern specification involved in mesonephros development(GO:0061227) renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) BMP signaling pathway involved in nephric duct formation(GO:0071893) glomerular visceral epithelial cell development(GO:0072015) comma-shaped body morphogenesis(GO:0072049) regulation of branch elongation involved in ureteric bud branching(GO:0072095) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in kidney development(GO:0072098) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) glomerulus morphogenesis(GO:0072102) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) mesenchymal cell proliferation involved in ureteric bud development(GO:0072138) nephric duct formation(GO:0072179) ureter urothelium development(GO:0072190) ureter epithelial cell differentiation(GO:0072192) ureter morphogenesis(GO:0072197) metanephric comma-shaped body morphogenesis(GO:0072278) glomerular epithelial cell development(GO:0072310) negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.4 |
1.3 |
GO:0042339 |
keratan sulfate metabolic process(GO:0042339) |
| 0.4 |
3.3 |
GO:0048251 |
elastic fiber assembly(GO:0048251) |
| 0.4 |
2.4 |
GO:0002036 |
regulation of L-glutamate transport(GO:0002036) |
| 0.4 |
1.9 |
GO:0048669 |
collateral sprouting in absence of injury(GO:0048669) |
| 0.4 |
2.2 |
GO:0032460 |
negative regulation of protein oligomerization(GO:0032460) negative regulation of protein homooligomerization(GO:0032463) |
| 0.4 |
1.1 |
GO:0002124 |
territorial aggressive behavior(GO:0002124) brainstem development(GO:0003360) vocalization behavior(GO:0071625) |
| 0.3 |
8.4 |
GO:0006884 |
cell volume homeostasis(GO:0006884) |
| 0.3 |
2.6 |
GO:0006689 |
ganglioside catabolic process(GO:0006689) |
| 0.3 |
3.6 |
GO:0051497 |
negative regulation of stress fiber assembly(GO:0051497) |
| 0.3 |
1.3 |
GO:0030913 |
paranodal junction assembly(GO:0030913) |
| 0.3 |
0.6 |
GO:0051014 |
actin filament severing(GO:0051014) |
| 0.3 |
0.9 |
GO:0003289 |
septum primum development(GO:0003284) atrial septum primum morphogenesis(GO:0003289) intestinal epithelial cell differentiation(GO:0060575) |
| 0.3 |
1.8 |
GO:0009450 |
gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.3 |
1.7 |
GO:0015811 |
sulfur amino acid transport(GO:0000101) L-cystine transport(GO:0015811) |
| 0.3 |
0.8 |
GO:0045656 |
negative regulation of monocyte differentiation(GO:0045656) |
| 0.2 |
1.3 |
GO:0010832 |
negative regulation of myotube differentiation(GO:0010832) |
| 0.2 |
0.7 |
GO:0019367 |
fatty acid elongation, saturated fatty acid(GO:0019367) |
| 0.2 |
2.0 |
GO:0018126 |
protein hydroxylation(GO:0018126) |
| 0.2 |
1.7 |
GO:0006072 |
glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.2 |
2.7 |
GO:1904893 |
negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.2 |
1.0 |
GO:0006930 |
substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.2 |
0.6 |
GO:0007500 |
mesodermal cell fate determination(GO:0007500) |
| 0.2 |
0.2 |
GO:0010982 |
regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.2 |
0.8 |
GO:0017014 |
protein nitrosylation(GO:0017014) peptidyl-cysteine S-nitrosylation(GO:0018119) |
| 0.2 |
1.1 |
GO:0060242 |
contact inhibition(GO:0060242) |
| 0.2 |
2.8 |
GO:0007567 |
parturition(GO:0007567) |
| 0.2 |
0.7 |
GO:0006556 |
S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.2 |
2.4 |
GO:0030854 |
positive regulation of granulocyte differentiation(GO:0030854) |
| 0.2 |
1.5 |
GO:0035434 |
copper ion transmembrane transport(GO:0035434) |
| 0.2 |
0.5 |
GO:0060596 |
mammary placode formation(GO:0060596) |
| 0.2 |
2.8 |
GO:0050812 |
acetyl-CoA biosynthetic process from pyruvate(GO:0006086) regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.2 |
0.5 |
GO:0051088 |
PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.2 |
2.4 |
GO:0042921 |
glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.2 |
1.1 |
GO:0033034 |
positive regulation of myeloid cell apoptotic process(GO:0033034) positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.2 |
0.6 |
GO:0019276 |
UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.2 |
3.2 |
GO:0000045 |
autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.1 |
0.4 |
GO:0006420 |
arginyl-tRNA aminoacylation(GO:0006420) |
| 0.1 |
0.6 |
GO:0010871 |
negative regulation of receptor biosynthetic process(GO:0010871) |
| 0.1 |
1.5 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 |
2.0 |
GO:0050872 |
white fat cell differentiation(GO:0050872) |
| 0.1 |
0.5 |
GO:0015868 |
purine ribonucleotide transport(GO:0015868) |
| 0.1 |
0.6 |
GO:0014850 |
response to muscle activity(GO:0014850) |
| 0.1 |
0.5 |
GO:0046778 |
modification by virus of host mRNA processing(GO:0046778) |
| 0.1 |
0.9 |
GO:0005981 |
regulation of glycogen catabolic process(GO:0005981) |
| 0.1 |
6.0 |
GO:0006914 |
autophagy(GO:0006914) |
| 0.1 |
1.1 |
GO:0034638 |
phosphatidylcholine catabolic process(GO:0034638) glycerophospholipid catabolic process(GO:0046475) |
| 0.1 |
0.1 |
GO:0021859 |
pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.1 |
0.6 |
GO:0009235 |
cobalamin metabolic process(GO:0009235) |
| 0.1 |
1.3 |
GO:0090263 |
positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.1 |
1.0 |
GO:0048625 |
myoblast fate commitment(GO:0048625) |
| 0.1 |
5.4 |
GO:0030837 |
negative regulation of actin filament polymerization(GO:0030837) |
| 0.1 |
0.9 |
GO:0019388 |
galactose catabolic process(GO:0019388) |
| 0.1 |
5.9 |
GO:0046324 |
regulation of glucose import(GO:0046324) |
| 0.1 |
1.7 |
GO:0006750 |
glutathione biosynthetic process(GO:0006750) |
| 0.1 |
0.9 |
GO:0032516 |
positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.1 |
0.3 |
GO:0019695 |
choline metabolic process(GO:0019695) |
| 0.1 |
0.8 |
GO:0006069 |
ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.1 |
0.7 |
GO:0060261 |
positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.1 |
0.1 |
GO:0006041 |
glucosamine metabolic process(GO:0006041) |
| 0.1 |
1.5 |
GO:0032410 |
negative regulation of transporter activity(GO:0032410) |
| 0.1 |
0.3 |
GO:0042727 |
flavin-containing compound biosynthetic process(GO:0042727) |
| 0.1 |
1.9 |
GO:0000038 |
very long-chain fatty acid metabolic process(GO:0000038) |
| 0.1 |
2.7 |
GO:0042147 |
retrograde transport, endosome to Golgi(GO:0042147) |
| 0.1 |
2.1 |
GO:0006957 |
complement activation, alternative pathway(GO:0006957) |
| 0.1 |
0.6 |
GO:2000172 |
regulation of branching morphogenesis of a nerve(GO:2000172) |
| 0.1 |
0.7 |
GO:0060009 |
Sertoli cell development(GO:0060009) |
| 0.1 |
0.5 |
GO:0032497 |
detection of lipopolysaccharide(GO:0032497) |
| 0.1 |
0.5 |
GO:0019852 |
L-ascorbic acid metabolic process(GO:0019852) |
| 0.1 |
0.3 |
GO:0006530 |
asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.1 |
2.0 |
GO:0080171 |
lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.1 |
0.2 |
GO:0006171 |
cAMP biosynthetic process(GO:0006171) |
| 0.1 |
1.3 |
GO:0045947 |
negative regulation of translational initiation(GO:0045947) |
| 0.1 |
1.3 |
GO:0033169 |
histone H3-K9 demethylation(GO:0033169) |
| 0.1 |
0.5 |
GO:0009750 |
response to fructose(GO:0009750) |
| 0.1 |
1.3 |
GO:0033574 |
response to testosterone(GO:0033574) |
| 0.1 |
0.2 |
GO:0050983 |
spermidine catabolic process(GO:0046203) deoxyhypusine biosynthetic process from spermidine(GO:0050983) |
| 0.1 |
1.6 |
GO:0032012 |
regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 |
0.5 |
GO:0040032 |
post-embryonic body morphogenesis(GO:0040032) |
| 0.1 |
0.3 |
GO:0030997 |
regulation of centriole-centriole cohesion(GO:0030997) |
| 0.1 |
0.9 |
GO:0000381 |
regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 |
0.7 |
GO:0007351 |
tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.1 |
0.3 |
GO:0043441 |
acetoacetic acid biosynthetic process(GO:0043441) |
| 0.1 |
1.4 |
GO:0009086 |
methionine biosynthetic process(GO:0009086) |
| 0.1 |
0.4 |
GO:0042693 |
muscle cell fate commitment(GO:0042693) |
| 0.1 |
0.3 |
GO:0035405 |
histone-threonine phosphorylation(GO:0035405) |
| 0.1 |
0.3 |
GO:0007492 |
endoderm development(GO:0007492) |
| 0.1 |
0.4 |
GO:0000244 |
spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 |
0.4 |
GO:0070245 |
positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.1 |
0.5 |
GO:0044341 |
sodium-dependent phosphate transport(GO:0044341) |
| 0.1 |
0.3 |
GO:0006874 |
cellular calcium ion homeostasis(GO:0006874) |
| 0.1 |
2.2 |
GO:0048286 |
lung alveolus development(GO:0048286) |
| 0.1 |
0.4 |
GO:0090342 |
regulation of cell aging(GO:0090342) |
| 0.1 |
1.0 |
GO:0006071 |
glycerol metabolic process(GO:0006071) |
| 0.1 |
1.5 |
GO:0003333 |
amino acid transmembrane transport(GO:0003333) |
| 0.1 |
0.8 |
GO:0008272 |
sulfate transport(GO:0008272) |
| 0.1 |
0.2 |
GO:0021570 |
rhombomere 4 development(GO:0021570) facial nucleus development(GO:0021754) |
| 0.1 |
1.1 |
GO:0071577 |
zinc II ion transmembrane transport(GO:0071577) |
| 0.1 |
0.2 |
GO:0032898 |
neurotrophin production(GO:0032898) |
| 0.1 |
0.9 |
GO:0008333 |
endosome to lysosome transport(GO:0008333) |
| 0.1 |
0.2 |
GO:0043418 |
cysteine biosynthetic process from serine(GO:0006535) homoserine metabolic process(GO:0009092) cysteine biosynthetic process via cystathionine(GO:0019343) transsulfuration(GO:0019346) homocysteine catabolic process(GO:0043418) |
| 0.1 |
0.9 |
GO:0007032 |
endosome organization(GO:0007032) |
| 0.1 |
1.5 |
GO:0006699 |
bile acid biosynthetic process(GO:0006699) |
| 0.1 |
1.6 |
GO:0051457 |
maintenance of protein location in nucleus(GO:0051457) |
| 0.1 |
0.1 |
GO:0002384 |
hepatic immune response(GO:0002384) |
| 0.1 |
6.6 |
GO:0018279 |
peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 |
2.3 |
GO:0006958 |
complement activation, classical pathway(GO:0006958) |
| 0.1 |
6.8 |
GO:0043281 |
regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043281) |
| 0.1 |
0.2 |
GO:1903825 |
organic acid transmembrane transport(GO:1903825) carboxylic acid transmembrane transport(GO:1905039) |
| 0.1 |
0.1 |
GO:0051928 |
positive regulation of calcium ion transport(GO:0051928) |
| 0.1 |
1.2 |
GO:0030199 |
collagen fibril organization(GO:0030199) |
| 0.1 |
0.2 |
GO:0050758 |
thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 |
0.2 |
GO:0043587 |
tongue morphogenesis(GO:0043587) |
| 0.0 |
0.8 |
GO:0032008 |
positive regulation of TOR signaling(GO:0032008) |
| 0.0 |
0.1 |
GO:0001732 |
formation of cytoplasmic translation initiation complex(GO:0001732) cytoplasmic translation(GO:0002181) cytoplasmic translational initiation(GO:0002183) |
| 0.0 |
0.1 |
GO:0019371 |
cyclooxygenase pathway(GO:0019371) |
| 0.0 |
0.1 |
GO:0032914 |
transforming growth factor beta1 production(GO:0032905) regulation of transforming growth factor beta1 production(GO:0032908) positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 |
0.3 |
GO:0070493 |
thrombin receptor signaling pathway(GO:0070493) |
| 0.0 |
2.0 |
GO:0071260 |
cellular response to mechanical stimulus(GO:0071260) |
| 0.0 |
1.0 |
GO:0048009 |
insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.0 |
1.7 |
GO:0051789 |
obsolete response to protein(GO:0051789) |
| 0.0 |
0.8 |
GO:0006656 |
phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 |
0.4 |
GO:0042354 |
fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 |
1.2 |
GO:0051225 |
spindle assembly(GO:0051225) |
| 0.0 |
0.3 |
GO:0046618 |
drug transmembrane transport(GO:0006855) drug export(GO:0046618) |
| 0.0 |
0.2 |
GO:0046835 |
carbohydrate phosphorylation(GO:0046835) |
| 0.0 |
1.1 |
GO:0030030 |
cell projection organization(GO:0030030) |
| 0.0 |
0.2 |
GO:0006564 |
L-serine biosynthetic process(GO:0006564) |
| 0.0 |
0.2 |
GO:0045767 |
obsolete regulation of anti-apoptosis(GO:0045767) |
| 0.0 |
0.9 |
GO:0046464 |
triglyceride catabolic process(GO:0019433) neutral lipid catabolic process(GO:0046461) acylglycerol catabolic process(GO:0046464) |
| 0.0 |
0.1 |
GO:1903301 |
lactate metabolic process(GO:0006089) regulation of glucokinase activity(GO:0033131) positive regulation of glucokinase activity(GO:0033133) regulation of hexokinase activity(GO:1903299) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 |
0.1 |
GO:0001973 |
adenosine receptor signaling pathway(GO:0001973) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 |
0.6 |
GO:0035036 |
binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) |
| 0.0 |
0.1 |
GO:0014887 |
muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 |
1.2 |
GO:0043484 |
regulation of RNA splicing(GO:0043484) |
| 0.0 |
0.0 |
GO:0051311 |
meiotic metaphase I plate congression(GO:0043060) meiotic metaphase plate congression(GO:0051311) |
| 0.0 |
0.3 |
GO:0071166 |
ribonucleoprotein complex localization(GO:0071166) ribonucleoprotein complex export from nucleus(GO:0071426) |
| 0.0 |
7.0 |
GO:0006869 |
lipid transport(GO:0006869) |
| 0.0 |
0.3 |
GO:0007217 |
tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 |
3.1 |
GO:0008277 |
regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 |
0.2 |
GO:0034287 |
type B pancreatic cell differentiation(GO:0003309) detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 |
0.1 |
GO:0018344 |
protein geranylgeranylation(GO:0018344) |
| 0.0 |
0.6 |
GO:0032570 |
response to progesterone(GO:0032570) |
| 0.0 |
0.3 |
GO:0060216 |
definitive hemopoiesis(GO:0060216) |
| 0.0 |
0.1 |
GO:0006051 |
mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.0 |
0.7 |
GO:0060334 |
regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 |
0.2 |
GO:0070327 |
thyroid hormone transport(GO:0070327) |
| 0.0 |
0.4 |
GO:0070389 |
chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 |
0.2 |
GO:0001782 |
B cell homeostasis(GO:0001782) |
| 0.0 |
0.1 |
GO:0007000 |
nucleolus organization(GO:0007000) |
| 0.0 |
0.3 |
GO:0050931 |
melanocyte differentiation(GO:0030318) pigment cell differentiation(GO:0050931) |
| 0.0 |
0.2 |
GO:0006931 |
substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 |
0.8 |
GO:0046847 |
filopodium assembly(GO:0046847) |
| 0.0 |
0.1 |
GO:0071539 |
protein localization to centrosome(GO:0071539) |
| 0.0 |
0.1 |
GO:0007616 |
long-term memory(GO:0007616) |
| 0.0 |
0.1 |
GO:0070407 |
oxidation-dependent protein catabolic process(GO:0070407) |
| 0.0 |
1.5 |
GO:0034976 |
response to endoplasmic reticulum stress(GO:0034976) |
| 0.0 |
0.4 |
GO:0015939 |
pantothenate metabolic process(GO:0015939) |
| 0.0 |
0.1 |
GO:0032747 |
positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 |
0.4 |
GO:0097031 |
NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 |
2.5 |
GO:0002576 |
platelet degranulation(GO:0002576) |
| 0.0 |
1.6 |
GO:0048706 |
embryonic skeletal system development(GO:0048706) |
| 0.0 |
0.2 |
GO:0007252 |
I-kappaB phosphorylation(GO:0007252) |
| 0.0 |
0.1 |
GO:0007197 |
adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 |
1.1 |
GO:0007569 |
cell aging(GO:0007569) |
| 0.0 |
0.2 |
GO:1904030 |
negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 |
1.0 |
GO:0006446 |
regulation of translational initiation(GO:0006446) |
| 0.0 |
0.1 |
GO:0034587 |
piRNA metabolic process(GO:0034587) |
| 0.0 |
0.0 |
GO:0060708 |
spongiotrophoblast differentiation(GO:0060708) |
| 0.0 |
0.1 |
GO:0000266 |
mitochondrial fission(GO:0000266) |
| 0.0 |
0.2 |
GO:0048199 |
vesicle targeting, to, from or within Golgi(GO:0048199) |
| 0.0 |
0.2 |
GO:0043687 |
post-translational protein modification(GO:0043687) |
| 0.0 |
0.1 |
GO:0000188 |
inactivation of MAPK activity(GO:0000188) |
| 0.0 |
0.1 |
GO:0072348 |
sulfur compound transport(GO:0072348) |
| 0.0 |
0.1 |
GO:0042417 |
dopamine metabolic process(GO:0042417) |
| 0.0 |
0.1 |
GO:0023041 |
neuronal signal transduction(GO:0023041) |
| 0.0 |
1.5 |
GO:0006368 |
transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 |
0.1 |
GO:0046697 |
decidualization(GO:0046697) |
| 0.0 |
0.8 |
GO:0007422 |
peripheral nervous system development(GO:0007422) |
| 0.0 |
2.0 |
GO:0006814 |
sodium ion transport(GO:0006814) |
| 0.0 |
0.6 |
GO:0014047 |
glutamate secretion(GO:0014047) |
| 0.0 |
0.3 |
GO:0005978 |
glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 |
0.3 |
GO:0072332 |
intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 |
0.0 |
GO:0007171 |
activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 |
0.6 |
GO:0048477 |
oogenesis(GO:0048477) |
| 0.0 |
0.1 |
GO:0051881 |
regulation of mitochondrial membrane potential(GO:0051881) |
| 0.0 |
0.4 |
GO:0042744 |
hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 |
0.1 |
GO:0018101 |
protein citrullination(GO:0018101) |
| 0.0 |
0.1 |
GO:0006104 |
succinyl-CoA metabolic process(GO:0006104) |
| 0.0 |
0.1 |
GO:0007586 |
digestion(GO:0007586) |
| 0.0 |
0.2 |
GO:0051297 |
centrosome organization(GO:0051297) |
| 0.0 |
0.2 |
GO:0040019 |
positive regulation of embryonic development(GO:0040019) |
| 0.0 |
0.1 |
GO:0048069 |
eye pigmentation(GO:0048069) |
| 0.0 |
0.3 |
GO:0005977 |
glycogen metabolic process(GO:0005977) |
| 0.0 |
0.7 |
GO:0000910 |
cytokinesis(GO:0000910) |
| 0.0 |
0.0 |
GO:0043084 |
penile erection(GO:0043084) |
| 0.0 |
0.1 |
GO:0071108 |
protein K48-linked deubiquitination(GO:0071108) |
| 0.0 |
0.3 |
GO:0045739 |
positive regulation of DNA repair(GO:0045739) |
| 0.0 |
0.0 |
GO:0018874 |
benzoate metabolic process(GO:0018874) |
| 0.0 |
0.2 |
GO:0006890 |
retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 |
0.2 |
GO:0035094 |
response to nicotine(GO:0035094) |
| 0.0 |
0.3 |
GO:0006939 |
smooth muscle contraction(GO:0006939) |
| 0.0 |
0.1 |
GO:0042149 |
cellular response to glucose starvation(GO:0042149) |
| 0.0 |
0.2 |
GO:0030815 |
negative regulation of cAMP metabolic process(GO:0030815) negative regulation of cAMP biosynthetic process(GO:0030818) |
| 0.0 |
3.0 |
GO:0007067 |
mitotic nuclear division(GO:0007067) |
| 0.0 |
0.1 |
GO:0006265 |
DNA topological change(GO:0006265) |
| 0.0 |
0.1 |
GO:0000186 |
activation of MAPKK activity(GO:0000186) |