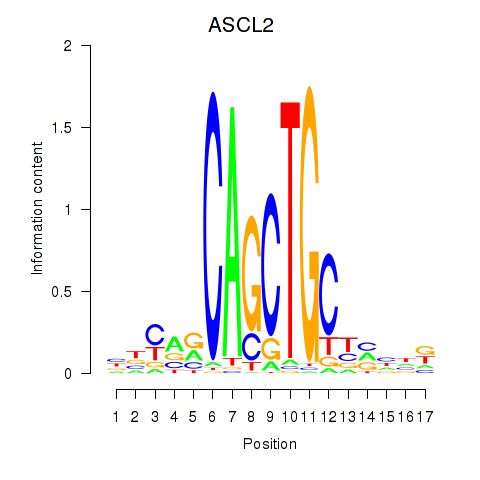
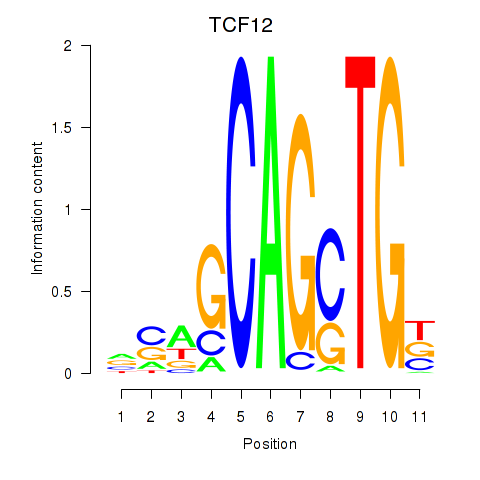
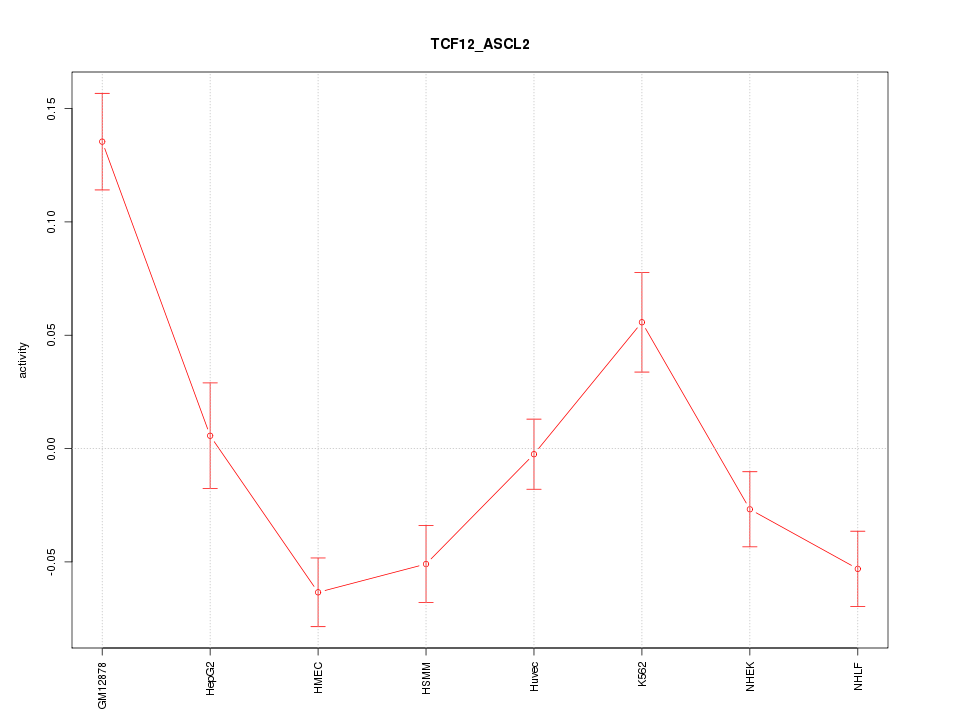
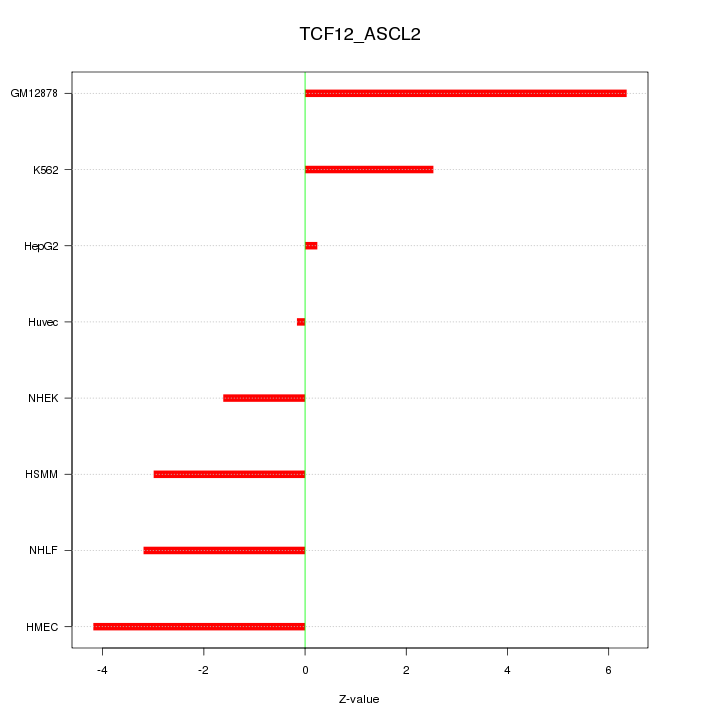
| 10.9 |
32.7 |
GO:0035408 |
histone H3-T6 phosphorylation(GO:0035408) |
| 5.6 |
16.9 |
GO:0000738 |
DNA catabolic process, exonucleolytic(GO:0000738) |
| 1.7 |
12.2 |
GO:0015755 |
fructose transport(GO:0015755) |
| 1.0 |
3.0 |
GO:0031558 |
obsolete induction of apoptosis in response to chemical stimulus(GO:0031558) |
| 0.8 |
4.2 |
GO:0008588 |
release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.8 |
2.5 |
GO:0042760 |
very long-chain fatty acid catabolic process(GO:0042760) |
| 0.7 |
2.1 |
GO:0002874 |
regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.7 |
2.0 |
GO:0051823 |
regulation of synapse structural plasticity(GO:0051823) |
| 0.7 |
4.6 |
GO:0045647 |
negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.7 |
2.6 |
GO:0021571 |
rhombomere 5 development(GO:0021571) |
| 0.6 |
0.6 |
GO:0021754 |
facial nucleus development(GO:0021754) |
| 0.6 |
5.4 |
GO:0045586 |
regulation of gamma-delta T cell differentiation(GO:0045586) positive regulation of gamma-delta T cell differentiation(GO:0045588) |
| 0.6 |
3.5 |
GO:0002430 |
complement receptor mediated signaling pathway(GO:0002430) |
| 0.6 |
1.7 |
GO:0048496 |
maintenance of organ identity(GO:0048496) |
| 0.6 |
1.7 |
GO:0001757 |
somite specification(GO:0001757) |
| 0.6 |
1.7 |
GO:0019886 |
antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.5 |
2.2 |
GO:0051534 |
negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.5 |
5.2 |
GO:0015074 |
DNA integration(GO:0015074) |
| 0.5 |
1.5 |
GO:0060318 |
definitive erythrocyte differentiation(GO:0060318) |
| 0.5 |
1.5 |
GO:0034126 |
regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) positive regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034126) |
| 0.5 |
1.4 |
GO:0002538 |
arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.5 |
3.3 |
GO:0090400 |
stress-induced premature senescence(GO:0090400) |
| 0.5 |
1.4 |
GO:0060316 |
positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.4 |
1.8 |
GO:0032929 |
negative regulation of superoxide anion generation(GO:0032929) |
| 0.4 |
2.2 |
GO:0072526 |
pyridine-containing compound catabolic process(GO:0072526) |
| 0.4 |
0.9 |
GO:2001259 |
positive regulation of calcium ion transmembrane transporter activity(GO:1901021) positive regulation of cation channel activity(GO:2001259) |
| 0.4 |
5.7 |
GO:0051489 |
regulation of filopodium assembly(GO:0051489) |
| 0.4 |
1.3 |
GO:0032328 |
alanine transport(GO:0032328) |
| 0.4 |
1.2 |
GO:0006167 |
AMP biosynthetic process(GO:0006167) |
| 0.4 |
2.3 |
GO:0009450 |
gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.4 |
1.9 |
GO:0003070 |
age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) |
| 0.4 |
4.0 |
GO:0006030 |
chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.4 |
1.4 |
GO:0035470 |
positive regulation of vascular wound healing(GO:0035470) |
| 0.4 |
1.4 |
GO:0033622 |
integrin activation(GO:0033622) regulation of integrin activation(GO:0033623) positive regulation of integrin activation(GO:0033625) |
| 0.4 |
1.4 |
GO:0051665 |
membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) membrane raft localization(GO:0051665) |
| 0.3 |
1.4 |
GO:0006768 |
biotin metabolic process(GO:0006768) |
| 0.3 |
1.0 |
GO:1903961 |
positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) positive regulation of anion transmembrane transport(GO:1903961) |
| 0.3 |
10.9 |
GO:0030851 |
granulocyte differentiation(GO:0030851) |
| 0.3 |
0.3 |
GO:0051147 |
regulation of muscle cell differentiation(GO:0051147) positive regulation of muscle cell differentiation(GO:0051149) |
| 0.3 |
8.3 |
GO:0050853 |
B cell receptor signaling pathway(GO:0050853) |
| 0.3 |
1.0 |
GO:0072677 |
eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.3 |
2.0 |
GO:0051051 |
negative regulation of transport(GO:0051051) |
| 0.3 |
2.6 |
GO:0033160 |
positive regulation of protein import into nucleus, translocation(GO:0033160) |
| 0.3 |
0.6 |
GO:0018149 |
peptide cross-linking(GO:0018149) |
| 0.3 |
1.0 |
GO:2000105 |
positive regulation of DNA-dependent DNA replication initiation(GO:0032298) positive regulation of DNA-dependent DNA replication(GO:2000105) |
| 0.3 |
1.3 |
GO:0015917 |
aminophospholipid transport(GO:0015917) |
| 0.3 |
1.6 |
GO:0006436 |
tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.3 |
0.6 |
GO:0035405 |
histone-threonine phosphorylation(GO:0035405) |
| 0.3 |
0.9 |
GO:0070171 |
negative regulation of tooth mineralization(GO:0070171) |
| 0.3 |
0.9 |
GO:0070340 |
detection of bacterial lipoprotein(GO:0042494) detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.3 |
0.9 |
GO:0002326 |
B cell lineage commitment(GO:0002326) |
| 0.3 |
1.5 |
GO:0009439 |
cyanate metabolic process(GO:0009439) cyanate catabolic process(GO:0009440) |
| 0.3 |
1.2 |
GO:0045199 |
maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.3 |
1.1 |
GO:0051573 |
negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.3 |
1.4 |
GO:0051781 |
positive regulation of cell division(GO:0051781) |
| 0.3 |
1.1 |
GO:0051964 |
negative regulation of synapse assembly(GO:0051964) |
| 0.3 |
1.1 |
GO:0002478 |
antigen processing and presentation of exogenous peptide antigen(GO:0002478) |
| 0.3 |
2.5 |
GO:1900087 |
traversing start control point of mitotic cell cycle(GO:0007089) positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) positive regulation of cell cycle G1/S phase transition(GO:1902808) |
| 0.3 |
2.2 |
GO:0046831 |
regulation of RNA export from nucleus(GO:0046831) |
| 0.3 |
0.8 |
GO:0031297 |
replication fork processing(GO:0031297) |
| 0.3 |
1.9 |
GO:0019264 |
glycine biosynthetic process from serine(GO:0019264) |
| 0.3 |
0.8 |
GO:0071280 |
cellular response to copper ion(GO:0071280) |
| 0.3 |
1.3 |
GO:0010508 |
positive regulation of autophagy(GO:0010508) |
| 0.3 |
0.8 |
GO:0007206 |
phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.3 |
0.8 |
GO:0040016 |
embryonic cleavage(GO:0040016) |
| 0.3 |
1.3 |
GO:0046952 |
ketone body catabolic process(GO:0046952) |
| 0.3 |
0.3 |
GO:0031018 |
endocrine pancreas development(GO:0031018) |
| 0.3 |
2.3 |
GO:0070327 |
thyroid hormone transport(GO:0070327) |
| 0.3 |
0.8 |
GO:0001807 |
type IV hypersensitivity(GO:0001806) regulation of type IV hypersensitivity(GO:0001807) |
| 0.2 |
1.0 |
GO:0045872 |
positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.2 |
1.2 |
GO:0072125 |
negative regulation of prostatic bud formation(GO:0060686) negative regulation of glomerular mesangial cell proliferation(GO:0072125) negative regulation of glomerulus development(GO:0090194) negative regulation of cell proliferation involved in kidney development(GO:1901723) |
| 0.2 |
2.2 |
GO:0048799 |
organ maturation(GO:0048799) bone maturation(GO:0070977) |
| 0.2 |
1.9 |
GO:0006561 |
proline biosynthetic process(GO:0006561) |
| 0.2 |
3.6 |
GO:0019221 |
cytokine-mediated signaling pathway(GO:0019221) |
| 0.2 |
0.7 |
GO:0010041 |
response to iron(III) ion(GO:0010041) positive regulation of histone phosphorylation(GO:0033129) response to DDT(GO:0046680) regulation of chromosome condensation(GO:0060623) cellular response to iron ion(GO:0071281) cellular response to iron(III) ion(GO:0071283) |
| 0.2 |
0.7 |
GO:0001705 |
ectoderm formation(GO:0001705) |
| 0.2 |
0.7 |
GO:0019521 |
aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.2 |
2.0 |
GO:0032464 |
positive regulation of protein homooligomerization(GO:0032464) |
| 0.2 |
1.1 |
GO:0032878 |
regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.2 |
0.5 |
GO:0033599 |
regulation of mammary gland epithelial cell proliferation(GO:0033599) |
| 0.2 |
0.2 |
GO:0071379 |
cellular response to prostaglandin stimulus(GO:0071379) |
| 0.2 |
1.3 |
GO:0010842 |
retina layer formation(GO:0010842) |
| 0.2 |
0.4 |
GO:0045992 |
negative regulation of embryonic development(GO:0045992) |
| 0.2 |
0.2 |
GO:0045618 |
positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.2 |
0.6 |
GO:0035623 |
renal glucose absorption(GO:0035623) |
| 0.2 |
0.6 |
GO:2000644 |
low-density lipoprotein particle receptor catabolic process(GO:0032802) regulation of low-density lipoprotein particle receptor catabolic process(GO:0032803) regulation of receptor catabolic process(GO:2000644) |
| 0.2 |
0.6 |
GO:0005997 |
xylulose metabolic process(GO:0005997) |
| 0.2 |
0.6 |
GO:0002637 |
regulation of immunoglobulin production(GO:0002637) positive regulation of immunoglobulin production(GO:0002639) |
| 0.2 |
0.8 |
GO:0001782 |
B cell homeostasis(GO:0001782) |
| 0.2 |
5.9 |
GO:0060334 |
regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.2 |
5.6 |
GO:0033209 |
tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.2 |
1.6 |
GO:0032836 |
glomerular basement membrane development(GO:0032836) |
| 0.2 |
1.5 |
GO:0007064 |
mitotic sister chromatid cohesion(GO:0007064) |
| 0.2 |
0.8 |
GO:0071168 |
negative regulation of sister chromatid cohesion(GO:0045875) protein localization to chromatin(GO:0071168) |
| 0.2 |
1.5 |
GO:0045084 |
positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.2 |
1.3 |
GO:0090080 |
positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.2 |
2.2 |
GO:0050718 |
positive regulation of interleukin-1 beta secretion(GO:0050718) |
| 0.2 |
0.9 |
GO:0072202 |
cell differentiation involved in metanephros development(GO:0072202) |
| 0.2 |
3.3 |
GO:0006386 |
transcription elongation from RNA polymerase III promoter(GO:0006385) termination of RNA polymerase III transcription(GO:0006386) |
| 0.2 |
4.0 |
GO:0002504 |
antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.2 |
0.5 |
GO:0006419 |
alanyl-tRNA aminoacylation(GO:0006419) |
| 0.2 |
0.7 |
GO:0030263 |
apoptotic chromosome condensation(GO:0030263) |
| 0.2 |
1.1 |
GO:0046599 |
regulation of centriole replication(GO:0046599) |
| 0.2 |
1.1 |
GO:0014820 |
tonic smooth muscle contraction(GO:0014820) artery smooth muscle contraction(GO:0014824) |
| 0.2 |
0.3 |
GO:0034135 |
regulation of toll-like receptor 2 signaling pathway(GO:0034135) |
| 0.2 |
0.2 |
GO:0051928 |
positive regulation of calcium ion transport(GO:0051928) |
| 0.2 |
0.5 |
GO:0070634 |
transepithelial ammonium transport(GO:0070634) |
| 0.2 |
0.3 |
GO:1902170 |
cellular response to nitric oxide(GO:0071732) cellular response to reactive nitrogen species(GO:1902170) |
| 0.2 |
1.3 |
GO:0060122 |
auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) inner ear receptor stereocilium organization(GO:0060122) |
| 0.2 |
0.7 |
GO:0009103 |
lipopolysaccharide biosynthetic process(GO:0009103) |
| 0.2 |
1.2 |
GO:0035024 |
negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.2 |
2.6 |
GO:0006004 |
fucose metabolic process(GO:0006004) |
| 0.2 |
0.5 |
GO:0070544 |
histone H3-K36 demethylation(GO:0070544) |
| 0.2 |
0.6 |
GO:0030889 |
negative regulation of B cell proliferation(GO:0030889) |
| 0.2 |
0.5 |
GO:0034498 |
early endosome to Golgi transport(GO:0034498) |
| 0.2 |
0.8 |
GO:0042637 |
catagen(GO:0042637) |
| 0.2 |
0.8 |
GO:0010447 |
response to acidic pH(GO:0010447) |
| 0.2 |
1.4 |
GO:0090399 |
replicative senescence(GO:0090399) |
| 0.2 |
1.4 |
GO:0070373 |
negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.2 |
0.5 |
GO:0046015 |
carbon catabolite regulation of transcription(GO:0045990) regulation of transcription by glucose(GO:0046015) positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.2 |
0.2 |
GO:0046449 |
allantoin metabolic process(GO:0000255) creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.2 |
1.2 |
GO:0015862 |
uridine transport(GO:0015862) |
| 0.1 |
0.4 |
GO:0048147 |
negative regulation of fibroblast proliferation(GO:0048147) |
| 0.1 |
3.1 |
GO:0030593 |
neutrophil chemotaxis(GO:0030593) neutrophil migration(GO:1990266) |
| 0.1 |
1.8 |
GO:0071479 |
cellular response to ionizing radiation(GO:0071479) |
| 0.1 |
1.3 |
GO:0032232 |
negative regulation of actin filament bundle assembly(GO:0032232) |
| 0.1 |
0.9 |
GO:0008089 |
anterograde axonal transport(GO:0008089) |
| 0.1 |
0.4 |
GO:0045578 |
negative regulation of B cell differentiation(GO:0045578) |
| 0.1 |
0.4 |
GO:0045603 |
positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.1 |
0.4 |
GO:0046210 |
nitric oxide catabolic process(GO:0046210) |
| 0.1 |
1.3 |
GO:0032647 |
interferon-alpha production(GO:0032607) regulation of interferon-alpha production(GO:0032647) positive regulation of interferon-alpha production(GO:0032727) |
| 0.1 |
0.4 |
GO:0048752 |
semicircular canal morphogenesis(GO:0048752) |
| 0.1 |
1.4 |
GO:0061756 |
leukocyte tethering or rolling(GO:0050901) leukocyte adhesion to vascular endothelial cell(GO:0061756) |
| 0.1 |
0.4 |
GO:0019918 |
peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.1 |
0.5 |
GO:0031117 |
positive regulation of microtubule depolymerization(GO:0031117) |
| 0.1 |
0.5 |
GO:0046340 |
diacylglycerol catabolic process(GO:0046340) |
| 0.1 |
1.2 |
GO:0046051 |
GTP biosynthetic process(GO:0006183) UTP biosynthetic process(GO:0006228) CTP biosynthetic process(GO:0006241) pyrimidine ribonucleoside triphosphate metabolic process(GO:0009208) pyrimidine ribonucleoside triphosphate biosynthetic process(GO:0009209) CTP metabolic process(GO:0046036) UTP metabolic process(GO:0046051) |
| 0.1 |
1.4 |
GO:0050858 |
negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 |
0.4 |
GO:0008050 |
female courtship behavior(GO:0008050) |
| 0.1 |
0.6 |
GO:0032020 |
ISG15-protein conjugation(GO:0032020) |
| 0.1 |
0.4 |
GO:0072216 |
positive regulation of metanephros development(GO:0072216) regulation of cell proliferation involved in kidney development(GO:1901722) positive regulation of cell proliferation involved in kidney development(GO:1901724) |
| 0.1 |
1.8 |
GO:0046580 |
negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.1 |
2.0 |
GO:0031648 |
protein destabilization(GO:0031648) |
| 0.1 |
1.5 |
GO:0060042 |
retina morphogenesis in camera-type eye(GO:0060042) |
| 0.1 |
0.5 |
GO:0015853 |
adenine transport(GO:0015853) |
| 0.1 |
1.1 |
GO:0046835 |
carbohydrate phosphorylation(GO:0046835) |
| 0.1 |
0.4 |
GO:0006420 |
arginyl-tRNA aminoacylation(GO:0006420) |
| 0.1 |
0.9 |
GO:0033261 |
obsolete regulation of S phase(GO:0033261) |
| 0.1 |
1.6 |
GO:0002566 |
somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.1 |
1.8 |
GO:0000090 |
mitotic anaphase(GO:0000090) |
| 0.1 |
5.5 |
GO:0019079 |
viral genome replication(GO:0019079) |
| 0.1 |
0.2 |
GO:0060398 |
regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.1 |
0.9 |
GO:0016075 |
rRNA catabolic process(GO:0016075) |
| 0.1 |
1.2 |
GO:0016180 |
snRNA processing(GO:0016180) |
| 0.1 |
0.1 |
GO:0032608 |
interferon-beta production(GO:0032608) regulation of interferon-beta production(GO:0032648) |
| 0.1 |
0.4 |
GO:1904396 |
regulation of synaptic growth at neuromuscular junction(GO:0008582) regulation of neuromuscular junction development(GO:1904396) |
| 0.1 |
1.0 |
GO:0043011 |
myeloid dendritic cell differentiation(GO:0043011) dendritic cell differentiation(GO:0097028) |
| 0.1 |
1.2 |
GO:0033600 |
negative regulation of mammary gland epithelial cell proliferation(GO:0033600) regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.1 |
0.2 |
GO:0003077 |
obsolete negative regulation of diuresis(GO:0003077) |
| 0.1 |
0.9 |
GO:0015705 |
iodide transport(GO:0015705) |
| 0.1 |
0.6 |
GO:0033326 |
cerebrospinal fluid secretion(GO:0033326) |
| 0.1 |
0.3 |
GO:0034145 |
positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) |
| 0.1 |
0.6 |
GO:0006975 |
DNA damage induced protein phosphorylation(GO:0006975) |
| 0.1 |
1.0 |
GO:0030147 |
obsolete natriuresis(GO:0030147) |
| 0.1 |
0.1 |
GO:0060535 |
trachea cartilage morphogenesis(GO:0060535) cartilage morphogenesis(GO:0060536) |
| 0.1 |
0.6 |
GO:0051481 |
negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.1 |
0.3 |
GO:0048642 |
negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.1 |
0.9 |
GO:0035095 |
behavioral response to nicotine(GO:0035095) |
| 0.1 |
0.2 |
GO:0014889 |
muscle atrophy(GO:0014889) |
| 0.1 |
0.5 |
GO:0016973 |
poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 |
0.8 |
GO:0046339 |
diacylglycerol metabolic process(GO:0046339) |
| 0.1 |
0.7 |
GO:0009744 |
response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.1 |
0.4 |
GO:0045945 |
positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.1 |
0.1 |
GO:0051582 |
positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.1 |
0.1 |
GO:0002679 |
respiratory burst involved in defense response(GO:0002679) |
| 0.1 |
0.5 |
GO:0043306 |
positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of leukocyte degranulation(GO:0043302) positive regulation of mast cell degranulation(GO:0043306) |
| 0.1 |
0.7 |
GO:0042769 |
DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 |
0.4 |
GO:0006384 |
transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 |
0.3 |
GO:0060259 |
regulation of feeding behavior(GO:0060259) |
| 0.1 |
0.3 |
GO:0007628 |
adult walking behavior(GO:0007628) walking behavior(GO:0090659) |
| 0.1 |
0.5 |
GO:0015693 |
magnesium ion transport(GO:0015693) |
| 0.1 |
0.3 |
GO:0010759 |
positive regulation of macrophage chemotaxis(GO:0010759) mononuclear cell migration(GO:0071674) regulation of mononuclear cell migration(GO:0071675) |
| 0.1 |
0.7 |
GO:0000012 |
single strand break repair(GO:0000012) |
| 0.1 |
0.5 |
GO:0031665 |
negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.1 |
0.5 |
GO:0007023 |
post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 |
0.6 |
GO:1901503 |
ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.1 |
0.3 |
GO:0014063 |
regulation of serotonin secretion(GO:0014062) negative regulation of serotonin secretion(GO:0014063) |
| 0.1 |
0.3 |
GO:0042560 |
10-formyltetrahydrofolate metabolic process(GO:0009256) 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.1 |
0.2 |
GO:0050849 |
negative regulation of calcium-mediated signaling(GO:0050849) |
| 0.1 |
0.1 |
GO:0046101 |
hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.1 |
0.6 |
GO:0002295 |
T-helper cell lineage commitment(GO:0002295) alpha-beta T cell lineage commitment(GO:0002363) CD4-positive, alpha-beta T cell lineage commitment(GO:0043373) memory T cell differentiation(GO:0043379) regulation of memory T cell differentiation(GO:0043380) positive regulation of memory T cell differentiation(GO:0043382) T-helper 17 type immune response(GO:0072538) T-helper 17 cell differentiation(GO:0072539) T-helper 17 cell lineage commitment(GO:0072540) regulation of T-helper 17 type immune response(GO:2000316) positive regulation of T-helper 17 type immune response(GO:2000318) regulation of T-helper 17 cell differentiation(GO:2000319) positive regulation of T-helper 17 cell differentiation(GO:2000321) regulation of T-helper 17 cell lineage commitment(GO:2000328) positive regulation of T-helper 17 cell lineage commitment(GO:2000330) |
| 0.1 |
1.0 |
GO:0045824 |
negative regulation of innate immune response(GO:0045824) |
| 0.1 |
3.3 |
GO:0070830 |
bicellular tight junction assembly(GO:0070830) |
| 0.1 |
0.5 |
GO:0030578 |
nuclear body organization(GO:0030575) PML body organization(GO:0030578) |
| 0.1 |
1.3 |
GO:0031998 |
regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.1 |
0.3 |
GO:0032074 |
negative regulation of nuclease activity(GO:0032074) |
| 0.1 |
1.6 |
GO:0042438 |
melanin metabolic process(GO:0006582) melanin biosynthetic process(GO:0042438) |
| 0.1 |
2.0 |
GO:0016578 |
histone deubiquitination(GO:0016578) |
| 0.1 |
0.3 |
GO:0090467 |
response to herbicide(GO:0009635) L-arginine import(GO:0043091) arginine import(GO:0090467) L-arginine transport(GO:1902023) |
| 0.1 |
0.4 |
GO:0021830 |
cerebral cortex tangential migration(GO:0021800) interneuron migration from the subpallium to the cortex(GO:0021830) cerebral cortex GABAergic interneuron migration(GO:0021853) cerebral cortex GABAergic interneuron development(GO:0021894) interneuron migration(GO:1904936) |
| 0.1 |
0.8 |
GO:0045003 |
DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.1 |
0.9 |
GO:0051568 |
histone H3-K4 methylation(GO:0051568) |
| 0.1 |
9.7 |
GO:0042113 |
B cell activation(GO:0042113) |
| 0.1 |
1.0 |
GO:0035590 |
purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.1 |
1.0 |
GO:0006929 |
substrate-dependent cell migration(GO:0006929) |
| 0.1 |
2.8 |
GO:0006099 |
tricarboxylic acid cycle(GO:0006099) |
| 0.1 |
0.8 |
GO:0015886 |
heme transport(GO:0015886) |
| 0.1 |
0.7 |
GO:0006177 |
GMP biosynthetic process(GO:0006177) |
| 0.1 |
0.3 |
GO:0010225 |
response to UV-C(GO:0010225) |
| 0.1 |
1.1 |
GO:0060674 |
placenta blood vessel development(GO:0060674) |
| 0.1 |
2.3 |
GO:0050830 |
defense response to Gram-positive bacterium(GO:0050830) |
| 0.1 |
0.4 |
GO:0006556 |
S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 |
0.3 |
GO:0071474 |
cellular water homeostasis(GO:0009992) cellular hyperosmotic response(GO:0071474) |
| 0.1 |
0.3 |
GO:0034332 |
adherens junction organization(GO:0034332) |
| 0.1 |
0.2 |
GO:0060061 |
Spemann organizer formation(GO:0060061) |
| 0.1 |
0.4 |
GO:0090267 |
positive regulation of spindle checkpoint(GO:0090232) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) positive regulation of cell cycle checkpoint(GO:1901978) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.1 |
0.5 |
GO:0007276 |
gamete generation(GO:0007276) |
| 0.1 |
0.4 |
GO:0021937 |
cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 0.1 |
0.4 |
GO:0043619 |
regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.1 |
1.2 |
GO:0045730 |
respiratory burst(GO:0045730) |
| 0.1 |
0.2 |
GO:0043049 |
otic placode formation(GO:0043049) |
| 0.1 |
1.0 |
GO:0070493 |
thrombin receptor signaling pathway(GO:0070493) |
| 0.1 |
0.8 |
GO:0006020 |
inositol metabolic process(GO:0006020) |
| 0.1 |
0.7 |
GO:0043496 |
regulation of protein homodimerization activity(GO:0043496) |
| 0.1 |
0.2 |
GO:0046668 |
regulation of retinal cell programmed cell death(GO:0046668) |
| 0.1 |
0.2 |
GO:0045056 |
transcytosis(GO:0045056) |
| 0.1 |
2.7 |
GO:0050684 |
regulation of mRNA processing(GO:0050684) regulation of mRNA metabolic process(GO:1903311) |
| 0.1 |
0.6 |
GO:0001843 |
neural tube closure(GO:0001843) primary neural tube formation(GO:0014020) tube closure(GO:0060606) |
| 0.1 |
2.5 |
GO:0034508 |
centromere complex assembly(GO:0034508) |
| 0.1 |
0.7 |
GO:0034755 |
iron ion transmembrane transport(GO:0034755) |
| 0.1 |
0.3 |
GO:0035608 |
protein deglutamylation(GO:0035608) |
| 0.1 |
0.9 |
GO:0032784 |
regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.1 |
0.2 |
GO:0015670 |
carbon dioxide transport(GO:0015670) |
| 0.1 |
0.7 |
GO:0070189 |
kynurenine metabolic process(GO:0070189) |
| 0.1 |
0.5 |
GO:0001889 |
liver development(GO:0001889) hepaticobiliary system development(GO:0061008) |
| 0.1 |
1.7 |
GO:0002026 |
regulation of the force of heart contraction(GO:0002026) |
| 0.1 |
0.3 |
GO:0060363 |
cranial suture morphogenesis(GO:0060363) craniofacial suture morphogenesis(GO:0097094) |
| 0.1 |
0.4 |
GO:0061003 |
positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.1 |
1.8 |
GO:0015721 |
bile acid and bile salt transport(GO:0015721) |
| 0.1 |
0.3 |
GO:0031936 |
negative regulation of chromatin silencing(GO:0031936) |
| 0.1 |
0.3 |
GO:0034723 |
DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 |
1.6 |
GO:0070542 |
response to fatty acid(GO:0070542) |
| 0.1 |
0.4 |
GO:0008295 |
spermidine biosynthetic process(GO:0008295) |
| 0.1 |
2.7 |
GO:0006271 |
DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.1 |
1.1 |
GO:1901663 |
quinone biosynthetic process(GO:1901663) |
| 0.1 |
1.2 |
GO:0007020 |
microtubule nucleation(GO:0007020) |
| 0.1 |
0.2 |
GO:0060135 |
maternal process involved in female pregnancy(GO:0060135) |
| 0.1 |
0.3 |
GO:0032776 |
DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.1 |
0.4 |
GO:0007494 |
midgut development(GO:0007494) |
| 0.1 |
0.4 |
GO:0009235 |
cobalamin metabolic process(GO:0009235) |
| 0.1 |
0.1 |
GO:0014887 |
muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.1 |
1.0 |
GO:0006596 |
polyamine biosynthetic process(GO:0006596) |
| 0.1 |
4.9 |
GO:0000079 |
regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.1 |
0.3 |
GO:0097061 |
dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.1 |
1.0 |
GO:0015939 |
pantothenate metabolic process(GO:0015939) |
| 0.1 |
0.3 |
GO:0046855 |
phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 |
0.6 |
GO:0016998 |
cell wall macromolecule catabolic process(GO:0016998) cell wall macromolecule metabolic process(GO:0044036) cell wall organization or biogenesis(GO:0071554) |
| 0.1 |
0.4 |
GO:1903035 |
negative regulation of response to wounding(GO:1903035) |
| 0.1 |
0.3 |
GO:0003097 |
renal water transport(GO:0003097) |
| 0.1 |
0.6 |
GO:0032148 |
activation of protein kinase B activity(GO:0032148) |
| 0.1 |
0.5 |
GO:0045898 |
regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.1 |
0.3 |
GO:0007182 |
common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.1 |
0.1 |
GO:2000078 |
regulation of type B pancreatic cell development(GO:2000074) positive regulation of type B pancreatic cell development(GO:2000078) |
| 0.1 |
1.0 |
GO:0006071 |
glycerol metabolic process(GO:0006071) |
| 0.1 |
0.3 |
GO:0018125 |
peptidyl-cysteine methylation(GO:0018125) |
| 0.1 |
1.1 |
GO:0006700 |
C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.1 |
0.5 |
GO:0015985 |
energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 |
0.3 |
GO:0048840 |
otolith development(GO:0048840) |
| 0.1 |
0.5 |
GO:0046500 |
S-adenosylmethionine metabolic process(GO:0046500) |
| 0.1 |
0.7 |
GO:0006776 |
vitamin A metabolic process(GO:0006776) |
| 0.1 |
1.1 |
GO:0006298 |
mismatch repair(GO:0006298) |
| 0.1 |
0.4 |
GO:0006734 |
NADH metabolic process(GO:0006734) |
| 0.1 |
0.3 |
GO:0006878 |
cellular copper ion homeostasis(GO:0006878) |
| 0.1 |
0.1 |
GO:0009113 |
purine nucleobase biosynthetic process(GO:0009113) |
| 0.1 |
0.2 |
GO:0006560 |
proline metabolic process(GO:0006560) |
| 0.1 |
0.4 |
GO:0007016 |
cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.1 |
0.1 |
GO:0006513 |
protein monoubiquitination(GO:0006513) |
| 0.1 |
0.4 |
GO:0033962 |
cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.1 |
0.4 |
GO:0006188 |
IMP biosynthetic process(GO:0006188) IMP metabolic process(GO:0046040) |
| 0.1 |
0.2 |
GO:0043461 |
proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting two-sector ATPase complex assembly(GO:0070071) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.1 |
0.2 |
GO:0061302 |
smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 |
0.4 |
GO:0016080 |
synaptic vesicle targeting(GO:0016080) |
| 0.1 |
0.4 |
GO:0071459 |
protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.1 |
0.3 |
GO:0018106 |
peptidyl-histidine phosphorylation(GO:0018106) |
| 0.1 |
0.3 |
GO:0007340 |
acrosome reaction(GO:0007340) |
| 0.1 |
0.2 |
GO:0003416 |
endochondral bone growth(GO:0003416) growth plate cartilage development(GO:0003417) bone growth(GO:0098868) |
| 0.1 |
0.8 |
GO:0030252 |
growth hormone secretion(GO:0030252) |
| 0.1 |
0.6 |
GO:0006098 |
pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.1 |
1.2 |
GO:0000291 |
nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.1 |
0.3 |
GO:0019321 |
pentose metabolic process(GO:0019321) |
| 0.1 |
0.2 |
GO:0010919 |
regulation of inositol phosphate biosynthetic process(GO:0010919) |
| 0.1 |
1.3 |
GO:0015804 |
neutral amino acid transport(GO:0015804) |
| 0.1 |
0.4 |
GO:0020027 |
hemoglobin metabolic process(GO:0020027) |
| 0.1 |
0.4 |
GO:0021511 |
spinal cord patterning(GO:0021511) |
| 0.1 |
0.5 |
GO:0051973 |
positive regulation of telomerase activity(GO:0051973) |
| 0.1 |
0.2 |
GO:0051967 |
negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 |
0.1 |
GO:0000045 |
autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.1 |
0.1 |
GO:0030432 |
peristalsis(GO:0030432) |
| 0.1 |
0.9 |
GO:0006198 |
cAMP catabolic process(GO:0006198) |
| 0.1 |
1.3 |
GO:0006362 |
transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.1 |
0.3 |
GO:0007288 |
sperm axoneme assembly(GO:0007288) |
| 0.1 |
0.2 |
GO:0006003 |
fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 |
0.2 |
GO:0003351 |
epithelial cilium movement(GO:0003351) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.1 |
0.2 |
GO:0048075 |
regulation of eye pigmentation(GO:0048073) positive regulation of eye pigmentation(GO:0048075) |
| 0.1 |
0.1 |
GO:0060896 |
neural plate anterior/posterior regionalization(GO:0021999) neural plate pattern specification(GO:0060896) neural plate regionalization(GO:0060897) |
| 0.1 |
1.8 |
GO:0007214 |
gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 |
0.6 |
GO:0042178 |
xenobiotic catabolic process(GO:0042178) |
| 0.1 |
0.3 |
GO:0034374 |
low-density lipoprotein particle remodeling(GO:0034374) |
| 0.1 |
0.3 |
GO:0030828 |
positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.1 |
0.4 |
GO:0006379 |
mRNA cleavage(GO:0006379) |
| 0.0 |
0.2 |
GO:0046037 |
GMP metabolic process(GO:0046037) |
| 0.0 |
0.4 |
GO:0006546 |
glycine catabolic process(GO:0006546) |
| 0.0 |
0.5 |
GO:0002286 |
T cell activation involved in immune response(GO:0002286) |
| 0.0 |
0.1 |
GO:0070842 |
aggresome assembly(GO:0070842) |
| 0.0 |
0.4 |
GO:0006670 |
sphingosine metabolic process(GO:0006670) diol metabolic process(GO:0034311) |
| 0.0 |
0.3 |
GO:0034638 |
phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 |
0.8 |
GO:0014037 |
Schwann cell differentiation(GO:0014037) |
| 0.0 |
0.1 |
GO:0019372 |
lipoxygenase pathway(GO:0019372) |
| 0.0 |
0.1 |
GO:0015851 |
nucleobase transport(GO:0015851) |
| 0.0 |
0.2 |
GO:0070995 |
NADPH oxidation(GO:0070995) |
| 0.0 |
0.8 |
GO:0006270 |
DNA replication initiation(GO:0006270) |
| 0.0 |
1.1 |
GO:0070664 |
negative regulation of mononuclear cell proliferation(GO:0032945) negative regulation of lymphocyte proliferation(GO:0050672) negative regulation of leukocyte proliferation(GO:0070664) |
| 0.0 |
0.2 |
GO:0032455 |
nerve growth factor processing(GO:0032455) |
| 0.0 |
2.7 |
GO:0019722 |
calcium-mediated signaling(GO:0019722) |
| 0.0 |
0.2 |
GO:0019509 |
L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 |
2.2 |
GO:0030217 |
T cell differentiation(GO:0030217) |
| 0.0 |
1.2 |
GO:0007052 |
mitotic spindle organization(GO:0007052) |
| 0.0 |
0.1 |
GO:0051892 |
negative regulation of heart induction by canonical Wnt signaling pathway(GO:0003136) negative regulation of cell fate specification(GO:0009996) negative regulation of cardioblast cell fate specification(GO:0009997) negative regulation of cell fate commitment(GO:0010454) cardioblast cell fate commitment(GO:0042684) cardioblast cell fate specification(GO:0042685) regulation of cardioblast cell fate specification(GO:0042686) negative regulation of cardioblast differentiation(GO:0051892) cardiac cell fate specification(GO:0060912) canonical Wnt signaling pathway involved in heart development(GO:0061316) negative regulation of heart induction(GO:1901320) negative regulation of cardiocyte differentiation(GO:1905208) regulation of cardiac cell fate specification(GO:2000043) negative regulation of cardiac cell fate specification(GO:2000044) |
| 0.0 |
0.1 |
GO:0098781 |
rRNA transcription(GO:0009303) ncRNA transcription(GO:0098781) |
| 0.0 |
0.3 |
GO:0046033 |
AMP metabolic process(GO:0046033) |
| 0.0 |
0.2 |
GO:0007386 |
compartment pattern specification(GO:0007386) |
| 0.0 |
0.3 |
GO:0050912 |
detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) detection of chemical stimulus involved in sensory perception of taste(GO:0050912) |
| 0.0 |
1.2 |
GO:0006383 |
transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 |
0.1 |
GO:0051083 |
'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 |
0.2 |
GO:0010957 |
negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 |
0.4 |
GO:0034383 |
positive regulation of cholesterol storage(GO:0010886) low-density lipoprotein particle clearance(GO:0034383) |
| 0.0 |
1.3 |
GO:0007051 |
spindle organization(GO:0007051) |
| 0.0 |
0.4 |
GO:0045668 |
negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 |
0.4 |
GO:0046683 |
response to organophosphorus(GO:0046683) response to cAMP(GO:0051591) |
| 0.0 |
0.3 |
GO:0006452 |
translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 |
0.1 |
GO:0071344 |
diphosphate metabolic process(GO:0071344) |
| 0.0 |
0.1 |
GO:0045717 |
negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 |
0.2 |
GO:0021854 |
hypothalamus development(GO:0021854) |
| 0.0 |
6.8 |
GO:0006813 |
potassium ion transport(GO:0006813) |
| 0.0 |
1.5 |
GO:0006289 |
nucleotide-excision repair(GO:0006289) |
| 0.0 |
0.2 |
GO:0000076 |
DNA replication checkpoint(GO:0000076) |
| 0.0 |
0.9 |
GO:0006691 |
leukotriene metabolic process(GO:0006691) |
| 0.0 |
0.0 |
GO:0050908 |
detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 |
0.2 |
GO:0030538 |
embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 |
2.0 |
GO:0006120 |
mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 |
0.2 |
GO:0045654 |
positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 |
0.1 |
GO:0060423 |
foregut regionalization(GO:0060423) lung field specification(GO:0060424) primary lung bud formation(GO:0060431) lung induction(GO:0060492) regulation of branching involved in lung morphogenesis(GO:0061046) positive regulation of branching involved in lung morphogenesis(GO:0061047) iris morphogenesis(GO:0061072) |
| 0.0 |
0.3 |
GO:0016556 |
mRNA modification(GO:0016556) |
| 0.0 |
1.2 |
GO:0008156 |
negative regulation of DNA replication(GO:0008156) |
| 0.0 |
0.0 |
GO:0002374 |
cytokine secretion involved in immune response(GO:0002374) negative regulation of production of molecular mediator of immune response(GO:0002701) negative regulation of cytokine production involved in immune response(GO:0002719) regulation of cytokine secretion involved in immune response(GO:0002739) negative regulation of cytokine secretion involved in immune response(GO:0002740) |
| 0.0 |
0.1 |
GO:0060192 |
negative regulation of lipoprotein lipase activity(GO:0051005) negative regulation of lipase activity(GO:0060192) |
| 0.0 |
3.5 |
GO:0008654 |
phospholipid biosynthetic process(GO:0008654) |
| 0.0 |
0.1 |
GO:0021978 |
telencephalon regionalization(GO:0021978) |
| 0.0 |
0.7 |
GO:0000060 |
protein import into nucleus, translocation(GO:0000060) |
| 0.0 |
0.3 |
GO:0051319 |
mitotic G2 phase(GO:0000085) G2 phase(GO:0051319) |
| 0.0 |
0.6 |
GO:0007158 |
neuron cell-cell adhesion(GO:0007158) |
| 0.0 |
0.2 |
GO:0046689 |
response to mercury ion(GO:0046689) |
| 0.0 |
0.2 |
GO:0019255 |
glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 |
0.1 |
GO:0000239 |
pachytene(GO:0000239) |
| 0.0 |
0.6 |
GO:0030178 |
negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 |
0.3 |
GO:0007162 |
negative regulation of cell adhesion(GO:0007162) |
| 0.0 |
0.3 |
GO:0045197 |
establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 |
1.0 |
GO:0070588 |
calcium ion transmembrane transport(GO:0070588) |
| 0.0 |
0.2 |
GO:0019883 |
antigen processing and presentation of endogenous antigen(GO:0019883) |
| 0.0 |
0.1 |
GO:0001778 |
plasma membrane repair(GO:0001778) |
| 0.0 |
0.1 |
GO:0072028 |
branching involved in ureteric bud morphogenesis(GO:0001658) ureteric bud morphogenesis(GO:0060675) renal tubule morphogenesis(GO:0061333) nephron morphogenesis(GO:0072028) nephron tubule morphogenesis(GO:0072078) nephron epithelium morphogenesis(GO:0072088) mesonephric tubule morphogenesis(GO:0072171) |
| 0.0 |
0.4 |
GO:0007346 |
regulation of mitotic cell cycle(GO:0007346) |
| 0.0 |
0.3 |
GO:0030225 |
macrophage differentiation(GO:0030225) |
| 0.0 |
0.2 |
GO:0006884 |
cell volume homeostasis(GO:0006884) |
| 0.0 |
1.0 |
GO:0007416 |
synapse assembly(GO:0007416) |
| 0.0 |
0.7 |
GO:0006471 |
protein ADP-ribosylation(GO:0006471) |
| 0.0 |
0.4 |
GO:0046464 |
triglyceride catabolic process(GO:0019433) neutral lipid catabolic process(GO:0046461) acylglycerol catabolic process(GO:0046464) glycerolipid catabolic process(GO:0046503) |
| 0.0 |
0.2 |
GO:0001522 |
pseudouridine synthesis(GO:0001522) |
| 0.0 |
0.4 |
GO:0046928 |
regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 |
0.1 |
GO:0006682 |
galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 |
0.1 |
GO:0045163 |
clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 |
0.1 |
GO:0006398 |
mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 |
0.5 |
GO:0006378 |
mRNA polyadenylation(GO:0006378) |
| 0.0 |
1.0 |
GO:0006672 |
ceramide metabolic process(GO:0006672) |
| 0.0 |
0.3 |
GO:0006818 |
hydrogen transport(GO:0006818) |
| 0.0 |
0.9 |
GO:0030239 |
myofibril assembly(GO:0030239) |
| 0.0 |
0.3 |
GO:0007498 |
mesoderm development(GO:0007498) |
| 0.0 |
0.1 |
GO:0051013 |
microtubule severing(GO:0051013) |
| 0.0 |
0.2 |
GO:0043537 |
negative regulation of blood vessel endothelial cell migration(GO:0043537) |
| 0.0 |
0.1 |
GO:0072111 |
cell proliferation involved in kidney development(GO:0072111) |
| 0.0 |
0.1 |
GO:0001672 |
regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 |
1.8 |
GO:0000236 |
mitotic prometaphase(GO:0000236) |
| 0.0 |
0.2 |
GO:0006600 |
creatine metabolic process(GO:0006600) |
| 0.0 |
0.5 |
GO:0019933 |
cAMP-mediated signaling(GO:0019933) |
| 0.0 |
2.7 |
GO:0006364 |
rRNA processing(GO:0006364) rRNA metabolic process(GO:0016072) |
| 0.0 |
0.3 |
GO:0006790 |
sulfur compound metabolic process(GO:0006790) |
| 0.0 |
0.1 |
GO:0015811 |
sulfur amino acid transport(GO:0000101) L-cystine transport(GO:0015811) |
| 0.0 |
0.3 |
GO:0015914 |
phospholipid transport(GO:0015914) |
| 0.0 |
0.3 |
GO:0033169 |
histone H3-K9 demethylation(GO:0033169) |
| 0.0 |
0.1 |
GO:0015871 |
choline transport(GO:0015871) |
| 0.0 |
0.0 |
GO:0000028 |
ribosomal small subunit assembly(GO:0000028) ribosomal small subunit biogenesis(GO:0042274) |
| 0.0 |
0.3 |
GO:0010921 |
regulation of phosphatase activity(GO:0010921) |
| 0.0 |
0.2 |
GO:0006465 |
signal peptide processing(GO:0006465) |
| 0.0 |
0.3 |
GO:0008625 |
extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) extrinsic apoptotic signaling pathway(GO:0097191) |
| 0.0 |
0.3 |
GO:0045646 |
regulation of erythrocyte differentiation(GO:0045646) |
| 0.0 |
0.1 |
GO:0090231 |
regulation of spindle checkpoint(GO:0090231) |
| 0.0 |
1.6 |
GO:0006879 |
cellular iron ion homeostasis(GO:0006879) |
| 0.0 |
1.7 |
GO:0022904 |
respiratory electron transport chain(GO:0022904) |
| 0.0 |
0.4 |
GO:0016226 |
iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 |
0.2 |
GO:0048387 |
negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 |
0.2 |
GO:0070555 |
response to interleukin-1(GO:0070555) |
| 0.0 |
0.8 |
GO:0043484 |
regulation of RNA splicing(GO:0043484) |
| 0.0 |
0.4 |
GO:0001954 |
positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 |
0.2 |
GO:0060678 |
dichotomous subdivision of terminal units involved in ureteric bud branching(GO:0060678) |
| 0.0 |
0.1 |
GO:0034379 |
very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.0 |
0.2 |
GO:0046146 |
tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 |
0.5 |
GO:0033014 |
porphyrin-containing compound biosynthetic process(GO:0006779) tetrapyrrole biosynthetic process(GO:0033014) |
| 0.0 |
0.2 |
GO:0015781 |
pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 |
0.1 |
GO:0035455 |
response to interferon-alpha(GO:0035455) |
| 0.0 |
0.1 |
GO:0050774 |
negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 |
0.9 |
GO:0006369 |
termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 |
0.2 |
GO:0060113 |
inner ear receptor cell differentiation(GO:0060113) |
| 0.0 |
0.3 |
GO:0035249 |
synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 |
0.4 |
GO:0060338 |
regulation of type I interferon-mediated signaling pathway(GO:0060338) |
| 0.0 |
0.1 |
GO:0006816 |
calcium ion transport(GO:0006816) |
| 0.0 |
0.4 |
GO:0048015 |
phosphatidylinositol-mediated signaling(GO:0048015) inositol lipid-mediated signaling(GO:0048017) |
| 0.0 |
0.1 |
GO:0031115 |
negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 |
0.1 |
GO:0048681 |
negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.0 |
0.1 |
GO:0006707 |
cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 |
5.1 |
GO:0050776 |
regulation of immune response(GO:0050776) |
| 0.0 |
0.7 |
GO:0051693 |
actin filament capping(GO:0051693) |
| 0.0 |
2.3 |
GO:0006333 |
chromatin assembly or disassembly(GO:0006333) |
| 0.0 |
0.2 |
GO:0050687 |
negative regulation of response to biotic stimulus(GO:0002832) negative regulation of defense response to virus(GO:0050687) |
| 0.0 |
0.2 |
GO:0006069 |
ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.0 |
0.2 |
GO:0031109 |
microtubule polymerization or depolymerization(GO:0031109) regulation of microtubule polymerization or depolymerization(GO:0031110) regulation of microtubule-based process(GO:0032886) regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 |
0.1 |
GO:0018206 |
peptidyl-methionine modification(GO:0018206) |
| 0.0 |
0.1 |
GO:0048488 |
synaptic vesicle recycling(GO:0036465) synaptic vesicle endocytosis(GO:0048488) clathrin-mediated endocytosis(GO:0072583) |
| 0.0 |
0.0 |
GO:0007171 |
activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 |
0.1 |
GO:0007270 |
neuron-neuron synaptic transmission(GO:0007270) |
| 0.0 |
2.7 |
GO:0043087 |
regulation of GTPase activity(GO:0043087) |
| 0.0 |
0.3 |
GO:0000387 |
spliceosomal snRNP assembly(GO:0000387) |
| 0.0 |
0.2 |
GO:0051703 |
social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 |
0.1 |
GO:0005978 |
glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 |
0.1 |
GO:0000042 |
protein targeting to Golgi(GO:0000042) establishment of protein localization to Golgi(GO:0072600) |
| 0.0 |
0.4 |
GO:0032012 |
regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 |
0.1 |
GO:0007258 |
JUN phosphorylation(GO:0007258) |
| 0.0 |
0.2 |
GO:0009086 |
methionine biosynthetic process(GO:0009086) |
| 0.0 |
0.4 |
GO:0016254 |
preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 |
0.3 |
GO:0006000 |
fructose metabolic process(GO:0006000) |
| 0.0 |
0.1 |
GO:1901224 |
activation of NF-kappaB-inducing kinase activity(GO:0007250) NIK/NF-kappaB signaling(GO:0038061) regulation of NIK/NF-kappaB signaling(GO:1901222) positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.0 |
0.3 |
GO:0050663 |
cytokine secretion(GO:0050663) |
| 0.0 |
0.3 |
GO:0046329 |
negative regulation of JNK cascade(GO:0046329) |
| 0.0 |
0.3 |
GO:0042462 |
photoreceptor cell development(GO:0042461) eye photoreceptor cell development(GO:0042462) |
| 0.0 |
0.2 |
GO:0042738 |
exogenous drug catabolic process(GO:0042738) |
| 0.0 |
0.2 |
GO:0000080 |
mitotic G1 phase(GO:0000080) |
| 0.0 |
0.1 |
GO:0046470 |
phosphatidylcholine metabolic process(GO:0046470) |
| 0.0 |
0.8 |
GO:0006958 |
complement activation, classical pathway(GO:0006958) |
| 0.0 |
1.3 |
GO:0006413 |
translational initiation(GO:0006413) |
| 0.0 |
0.2 |
GO:0070389 |
chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 |
0.0 |
GO:0043569 |
negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 |
0.1 |
GO:0006687 |
glycosphingolipid metabolic process(GO:0006687) |
| 0.0 |
0.2 |
GO:0051865 |
protein autoubiquitination(GO:0051865) |
| 0.0 |
0.1 |
GO:0006477 |
protein sulfation(GO:0006477) |
| 0.0 |
0.0 |
GO:0035330 |
regulation of hippo signaling(GO:0035330) |
| 0.0 |
0.1 |
GO:0035025 |
positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 |
0.1 |
GO:0006826 |
iron ion transport(GO:0006826) |
| 0.0 |
0.9 |
GO:0007605 |
sensory perception of sound(GO:0007605) |
| 0.0 |
0.6 |
GO:0006338 |
chromatin remodeling(GO:0006338) |
| 0.0 |
2.6 |
GO:0006281 |
DNA repair(GO:0006281) |
| 0.0 |
0.3 |
GO:0036075 |
endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 |
0.2 |
GO:0030317 |
sperm motility(GO:0030317) |
| 0.0 |
0.1 |
GO:0033574 |
response to testosterone(GO:0033574) |
| 0.0 |
0.6 |
GO:0016055 |
Wnt signaling pathway(GO:0016055) cell-cell signaling by wnt(GO:0198738) |
| 0.0 |
0.5 |
GO:0022900 |
electron transport chain(GO:0022900) |
| 0.0 |
0.6 |
GO:0045666 |
positive regulation of neuron differentiation(GO:0045666) |
| 0.0 |
0.1 |
GO:0001578 |
microtubule bundle formation(GO:0001578) |
| 0.0 |
0.2 |
GO:0000209 |
protein polyubiquitination(GO:0000209) |
| 0.0 |
0.1 |
GO:0030010 |
establishment of cell polarity(GO:0030010) |
| 0.0 |
0.5 |
GO:0048839 |
inner ear development(GO:0048839) |
| 0.0 |
0.1 |
GO:0048268 |
clathrin coat assembly(GO:0048268) |
| 0.0 |
0.2 |
GO:0046627 |
negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.0 |
0.1 |
GO:1903729 |
regulation of establishment of protein localization to plasma membrane(GO:0090003) regulation of protein localization to plasma membrane(GO:1903076) regulation of plasma membrane organization(GO:1903729) regulation of protein localization to cell periphery(GO:1904375) |
| 0.0 |
0.1 |
GO:0060644 |
mammary gland epithelial cell differentiation(GO:0060644) |
| 0.0 |
0.1 |
GO:0007168 |
receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 |
0.0 |
GO:0006105 |
succinate metabolic process(GO:0006105) |
| 0.0 |
2.3 |
GO:0016568 |
chromatin modification(GO:0016568) |
| 0.0 |
0.2 |
GO:0009312 |
oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 |
0.5 |
GO:0000910 |
cytokinesis(GO:0000910) |
| 0.0 |
0.7 |
GO:0050796 |
regulation of insulin secretion(GO:0050796) |
| 0.0 |
2.3 |
GO:0006954 |
inflammatory response(GO:0006954) |
| 0.0 |
0.0 |
GO:0045494 |
photoreceptor cell maintenance(GO:0045494) |
| 0.0 |
0.3 |
GO:0009791 |
post-embryonic development(GO:0009791) |