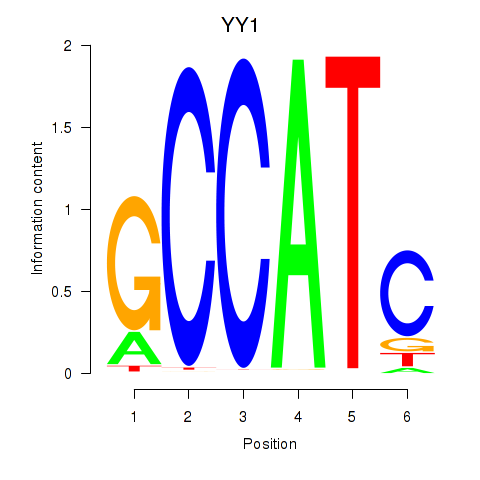
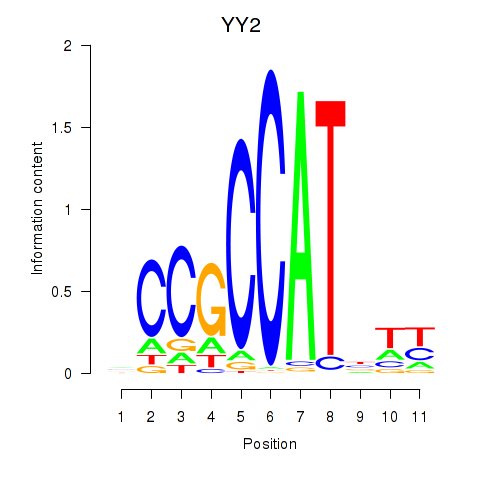
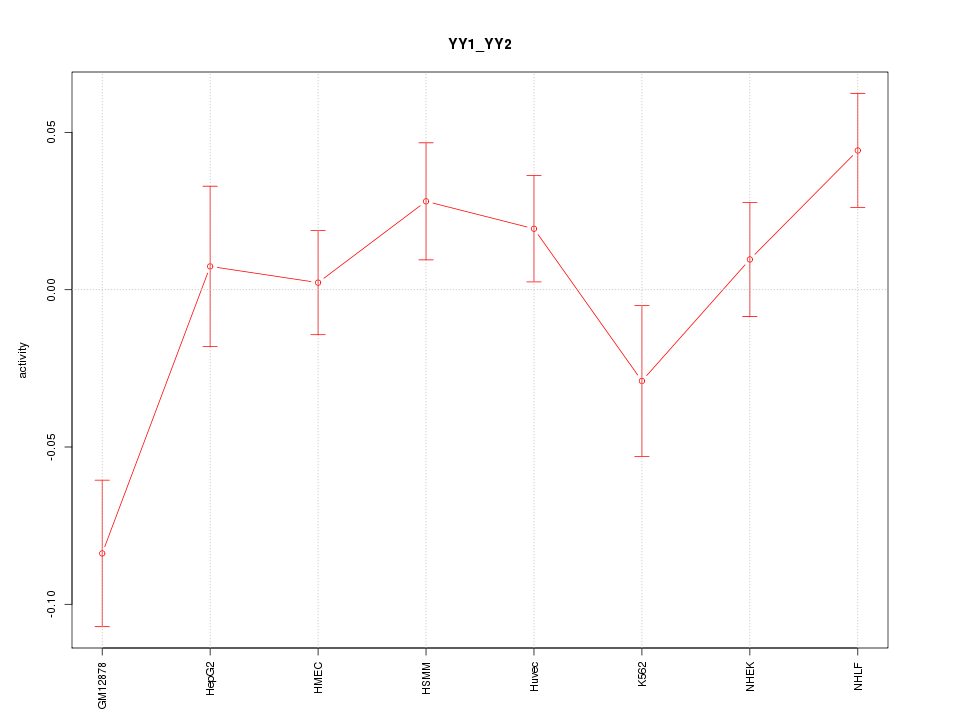
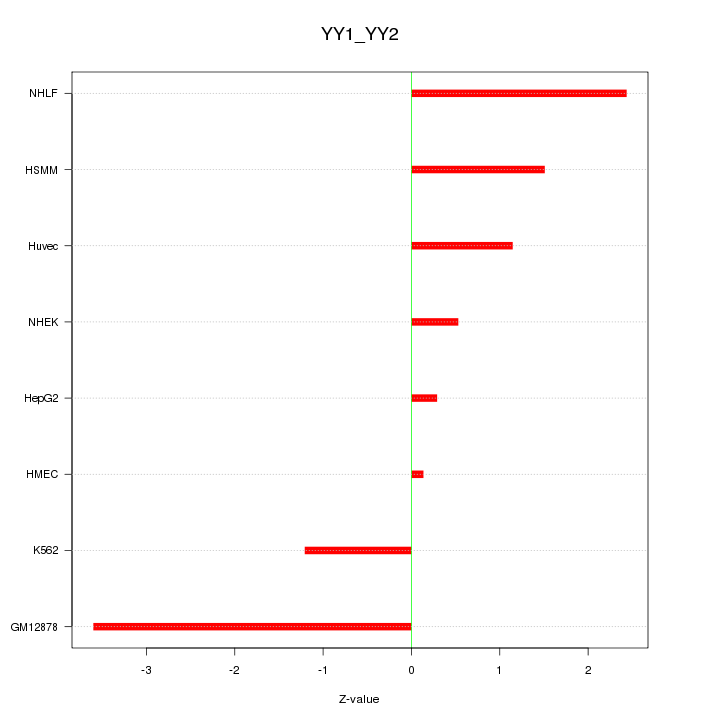
| 1.4 |
10.9 |
GO:0003065 |
regulation of heart rate by chemical signal(GO:0003062) positive regulation of heart rate by epinephrine(GO:0003065) |
| 1.1 |
4.4 |
GO:0019046 |
release from viral latency(GO:0019046) |
| 0.6 |
3.9 |
GO:0002036 |
regulation of L-glutamate transport(GO:0002036) |
| 0.6 |
2.2 |
GO:2000008 |
regulation of protein localization to cell surface(GO:2000008) |
| 0.5 |
2.1 |
GO:0002689 |
negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.5 |
1.5 |
GO:0019747 |
regulation of isoprenoid metabolic process(GO:0019747) |
| 0.5 |
0.9 |
GO:0045715 |
negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.5 |
1.4 |
GO:1904358 |
positive regulation of telomere maintenance via telomerase(GO:0032212) positive regulation of telomere maintenance via telomere lengthening(GO:1904358) |
| 0.4 |
1.3 |
GO:0006015 |
5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.4 |
1.1 |
GO:0070886 |
positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.3 |
0.3 |
GO:0032119 |
sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.3 |
0.9 |
GO:0060279 |
regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.3 |
1.2 |
GO:0033197 |
response to vitamin E(GO:0033197) |
| 0.3 |
3.4 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.3 |
0.3 |
GO:0051170 |
nuclear import(GO:0051170) |
| 0.3 |
1.2 |
GO:0051503 |
purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) |
| 0.3 |
2.3 |
GO:0032516 |
positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.3 |
1.3 |
GO:0050703 |
interleukin-1 alpha secretion(GO:0050703) |
| 0.3 |
0.8 |
GO:0019482 |
beta-alanine metabolic process(GO:0019482) |
| 0.2 |
0.7 |
GO:0009996 |
negative regulation of cell fate specification(GO:0009996) positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.2 |
0.7 |
GO:0097709 |
connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.2 |
1.4 |
GO:0014049 |
positive regulation of glutamate secretion(GO:0014049) |
| 0.2 |
2.3 |
GO:0010739 |
positive regulation of protein kinase A signaling(GO:0010739) |
| 0.2 |
1.4 |
GO:0014012 |
peripheral nervous system axon regeneration(GO:0014012) |
| 0.2 |
0.9 |
GO:0030913 |
paranodal junction assembly(GO:0030913) |
| 0.2 |
0.7 |
GO:0090047 |
obsolete positive regulation of transcription regulator activity(GO:0090047) |
| 0.2 |
0.9 |
GO:0072180 |
mesonephric duct morphogenesis(GO:0072180) |
| 0.2 |
0.7 |
GO:0035330 |
regulation of hippo signaling(GO:0035330) |
| 0.2 |
2.2 |
GO:0001967 |
suckling behavior(GO:0001967) |
| 0.2 |
0.6 |
GO:0060831 |
smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.2 |
0.6 |
GO:0006348 |
chromatin silencing at telomere(GO:0006348) |
| 0.2 |
0.6 |
GO:0018377 |
N-terminal protein myristoylation(GO:0006499) protein myristoylation(GO:0018377) |
| 0.2 |
0.6 |
GO:0032707 |
negative regulation of interleukin-23 production(GO:0032707) |
| 0.2 |
2.2 |
GO:0006621 |
protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.2 |
0.2 |
GO:0001816 |
cytokine production(GO:0001816) |
| 0.2 |
0.6 |
GO:0048549 |
positive regulation of pinocytosis(GO:0048549) |
| 0.2 |
0.5 |
GO:0007028 |
cytoplasm organization(GO:0007028) ribonucleoprotein complex disassembly(GO:0032988) |
| 0.2 |
0.9 |
GO:0001661 |
conditioned taste aversion(GO:0001661) |
| 0.2 |
2.0 |
GO:0060216 |
definitive hemopoiesis(GO:0060216) |
| 0.2 |
0.5 |
GO:0006427 |
histidyl-tRNA aminoacylation(GO:0006427) |
| 0.2 |
0.5 |
GO:0035120 |
post-embryonic appendage morphogenesis(GO:0035120) |
| 0.2 |
0.2 |
GO:0009101 |
glycoprotein biosynthetic process(GO:0009101) |
| 0.2 |
0.3 |
GO:0002082 |
regulation of oxidative phosphorylation(GO:0002082) |
| 0.2 |
1.1 |
GO:0030007 |
cellular potassium ion homeostasis(GO:0030007) negative regulation of cell volume(GO:0045794) |
| 0.2 |
0.5 |
GO:0045838 |
positive regulation of membrane potential(GO:0045838) |
| 0.2 |
0.5 |
GO:0016476 |
regulation of embryonic cell shape(GO:0016476) |
| 0.2 |
1.4 |
GO:0071692 |
sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.2 |
0.3 |
GO:0031133 |
regulation of axon diameter(GO:0031133) regulation of cell projection size(GO:0032536) |
| 0.2 |
0.2 |
GO:0043113 |
receptor clustering(GO:0043113) |
| 0.1 |
2.1 |
GO:0034446 |
substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.1 |
0.6 |
GO:0060209 |
estrus(GO:0060209) |
| 0.1 |
14.0 |
GO:0008064 |
regulation of actin polymerization or depolymerization(GO:0008064) |
| 0.1 |
0.4 |
GO:0035269 |
protein mannosylation(GO:0035268) protein O-linked mannosylation(GO:0035269) mannosylation(GO:0097502) |
| 0.1 |
0.4 |
GO:0003011 |
diaphragm contraction(GO:0002086) involuntary skeletal muscle contraction(GO:0003011) |
| 0.1 |
3.7 |
GO:0030199 |
collagen fibril organization(GO:0030199) |
| 0.1 |
0.5 |
GO:0001780 |
neutrophil homeostasis(GO:0001780) |
| 0.1 |
0.5 |
GO:0045218 |
zonula adherens maintenance(GO:0045218) |
| 0.1 |
0.7 |
GO:0048550 |
negative regulation of pinocytosis(GO:0048550) |
| 0.1 |
0.5 |
GO:0010716 |
regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.1 |
0.6 |
GO:0090343 |
positive regulation of cell aging(GO:0090343) |
| 0.1 |
0.8 |
GO:0046541 |
saliva secretion(GO:0046541) |
| 0.1 |
1.2 |
GO:0009650 |
UV protection(GO:0009650) |
| 0.1 |
0.1 |
GO:0007197 |
adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.1 |
0.5 |
GO:0030035 |
microspike assembly(GO:0030035) |
| 0.1 |
0.4 |
GO:0060911 |
cardiac cell fate commitment(GO:0060911) |
| 0.1 |
0.7 |
GO:0032007 |
negative regulation of TOR signaling(GO:0032007) |
| 0.1 |
0.5 |
GO:0009109 |
coenzyme catabolic process(GO:0009109) |
| 0.1 |
0.1 |
GO:0021543 |
pallium development(GO:0021543) |
| 0.1 |
3.0 |
GO:0006829 |
zinc II ion transport(GO:0006829) |
| 0.1 |
0.6 |
GO:0035020 |
regulation of Rac protein signal transduction(GO:0035020) |
| 0.1 |
0.3 |
GO:0007000 |
nucleolus organization(GO:0007000) |
| 0.1 |
2.0 |
GO:0043114 |
regulation of vascular permeability(GO:0043114) |
| 0.1 |
0.4 |
GO:0001824 |
blastocyst development(GO:0001824) |
| 0.1 |
0.9 |
GO:0006654 |
phosphatidic acid biosynthetic process(GO:0006654) |
| 0.1 |
1.0 |
GO:0007351 |
tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.1 |
0.3 |
GO:0042759 |
fatty acid elongation, saturated fatty acid(GO:0019367) long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 |
1.1 |
GO:0035308 |
negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.1 |
0.5 |
GO:0018344 |
protein geranylgeranylation(GO:0018344) |
| 0.1 |
0.3 |
GO:0002887 |
negative regulation of myeloid leukocyte mediated immunity(GO:0002887) negative regulation of leukocyte degranulation(GO:0043301) establishment of epithelial cell apical/basal polarity(GO:0045198) epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.1 |
0.3 |
GO:0046010 |
positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) |
| 0.1 |
0.4 |
GO:0042271 |
susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.1 |
1.0 |
GO:0060391 |
positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.1 |
2.6 |
GO:0031581 |
hemidesmosome assembly(GO:0031581) |
| 0.1 |
0.3 |
GO:0010248 |
establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.1 |
1.4 |
GO:0007032 |
endosome organization(GO:0007032) |
| 0.1 |
3.4 |
GO:0007041 |
lysosomal transport(GO:0007041) |
| 0.1 |
0.1 |
GO:0051092 |
positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.1 |
0.3 |
GO:0046477 |
glycosylceramide catabolic process(GO:0046477) |
| 0.1 |
0.9 |
GO:0050435 |
beta-amyloid metabolic process(GO:0050435) |
| 0.1 |
0.3 |
GO:0071418 |
cellular response to amine stimulus(GO:0071418) |
| 0.1 |
0.7 |
GO:0044341 |
sodium-dependent phosphate transport(GO:0044341) |
| 0.1 |
0.2 |
GO:0043461 |
proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting two-sector ATPase complex assembly(GO:0070071) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.1 |
0.7 |
GO:0060666 |
dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 |
0.6 |
GO:0032922 |
circadian regulation of gene expression(GO:0032922) |
| 0.1 |
0.5 |
GO:0043249 |
erythrocyte maturation(GO:0043249) |
| 0.1 |
0.3 |
GO:0042518 |
negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.1 |
0.6 |
GO:0071786 |
endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 |
0.4 |
GO:0006880 |
intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 |
0.2 |
GO:0070672 |
response to interleukin-15(GO:0070672) |
| 0.1 |
0.1 |
GO:0010834 |
obsolete telomere maintenance via telomere shortening(GO:0010834) |
| 0.1 |
0.4 |
GO:0070682 |
proteasome regulatory particle assembly(GO:0070682) |
| 0.1 |
0.6 |
GO:0035162 |
embryonic hemopoiesis(GO:0035162) |
| 0.1 |
0.1 |
GO:1903306 |
negative regulation of regulated secretory pathway(GO:1903306) |
| 0.1 |
0.3 |
GO:0043441 |
acetoacetic acid biosynthetic process(GO:0043441) |
| 0.1 |
0.1 |
GO:0032525 |
somite rostral/caudal axis specification(GO:0032525) |
| 0.1 |
0.7 |
GO:0051291 |
protein heterooligomerization(GO:0051291) |
| 0.1 |
0.2 |
GO:0007256 |
activation of JNKK activity(GO:0007256) |
| 0.1 |
0.5 |
GO:0006013 |
mannose metabolic process(GO:0006013) |
| 0.1 |
0.3 |
GO:0003322 |
pancreatic A cell development(GO:0003322) |
| 0.1 |
0.4 |
GO:0016255 |
attachment of GPI anchor to protein(GO:0016255) |
| 0.1 |
0.6 |
GO:0060158 |
phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 |
0.3 |
GO:0016236 |
macroautophagy(GO:0016236) |
| 0.1 |
0.2 |
GO:0048821 |
erythrocyte development(GO:0048821) myeloid cell development(GO:0061515) |
| 0.1 |
0.6 |
GO:0000209 |
protein polyubiquitination(GO:0000209) |
| 0.1 |
0.2 |
GO:0090136 |
epithelial cell-cell adhesion(GO:0090136) |
| 0.1 |
0.2 |
GO:0060206 |
estrous cycle phase(GO:0060206) |
| 0.1 |
0.7 |
GO:0051085 |
chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 |
0.2 |
GO:0034086 |
maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.1 |
0.6 |
GO:0045879 |
negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.1 |
0.2 |
GO:0061188 |
regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.1 |
0.3 |
GO:0034063 |
stress granule assembly(GO:0034063) |
| 0.1 |
0.7 |
GO:0048261 |
negative regulation of receptor-mediated endocytosis(GO:0048261) |
| 0.1 |
0.5 |
GO:0006685 |
sphingomyelin catabolic process(GO:0006685) |
| 0.1 |
4.7 |
GO:0008360 |
regulation of cell shape(GO:0008360) |
| 0.1 |
2.2 |
GO:0030520 |
intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.1 |
0.8 |
GO:0030853 |
negative regulation of granulocyte differentiation(GO:0030853) |
| 0.1 |
1.0 |
GO:0051131 |
chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 |
0.3 |
GO:0002051 |
osteoblast fate commitment(GO:0002051) |
| 0.1 |
0.4 |
GO:0042693 |
muscle cell fate commitment(GO:0042693) |
| 0.1 |
0.2 |
GO:0034551 |
respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 |
0.4 |
GO:0019388 |
galactose catabolic process(GO:0019388) |
| 0.1 |
0.6 |
GO:0016601 |
Rac protein signal transduction(GO:0016601) |
| 0.1 |
0.1 |
GO:0007403 |
glial cell fate determination(GO:0007403) |
| 0.1 |
0.3 |
GO:0030263 |
apoptotic chromosome condensation(GO:0030263) |
| 0.1 |
1.2 |
GO:0080171 |
lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.1 |
1.3 |
GO:0006490 |
dolichol-linked oligosaccharide biosynthetic process(GO:0006488) oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 |
0.3 |
GO:0007289 |
spermatid nucleus differentiation(GO:0007289) |
| 0.0 |
0.1 |
GO:0030997 |
regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 |
0.4 |
GO:0042987 |
amyloid precursor protein catabolic process(GO:0042987) |
| 0.0 |
1.4 |
GO:0031532 |
actin cytoskeleton reorganization(GO:0031532) |
| 0.0 |
0.1 |
GO:0006448 |
regulation of translational elongation(GO:0006448) |
| 0.0 |
0.5 |
GO:0033631 |
cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 |
0.9 |
GO:0035303 |
regulation of dephosphorylation(GO:0035303) |
| 0.0 |
3.4 |
GO:0030049 |
muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) actin-mediated cell contraction(GO:0070252) |
| 0.0 |
0.3 |
GO:0045916 |
negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 |
0.3 |
GO:0035335 |
peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 |
0.3 |
GO:0009136 |
ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 0.0 |
0.3 |
GO:0032968 |
positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 |
0.1 |
GO:0032571 |
response to vitamin K(GO:0032571) |
| 0.0 |
0.2 |
GO:0055009 |
atrial cardiac muscle tissue development(GO:0003228) atrial cardiac muscle tissue morphogenesis(GO:0055009) foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) epithelial cell proliferation involved in lung morphogenesis(GO:0060502) regulation of branching involved in lung morphogenesis(GO:0061046) positive regulation of branching involved in lung morphogenesis(GO:0061047) iris morphogenesis(GO:0061072) regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000794) |
| 0.0 |
0.0 |
GO:0010157 |
response to chlorate(GO:0010157) |
| 0.0 |
0.3 |
GO:0043045 |
DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.0 |
0.2 |
GO:0071108 |
protein K48-linked deubiquitination(GO:0071108) |
| 0.0 |
1.2 |
GO:0015914 |
phospholipid transport(GO:0015914) |
| 0.0 |
0.1 |
GO:0021801 |
cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.0 |
0.2 |
GO:0050668 |
positive regulation of sulfur amino acid metabolic process(GO:0031337) negative regulation of cellular amine metabolic process(GO:0033239) positive regulation of cellular amino acid metabolic process(GO:0045764) negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) positive regulation of homocysteine metabolic process(GO:0050668) |
| 0.0 |
0.1 |
GO:0036315 |
cellular response to sterol(GO:0036315) cellular response to cholesterol(GO:0071397) |
| 0.0 |
0.2 |
GO:0010919 |
regulation of inositol phosphate biosynthetic process(GO:0010919) positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 |
0.1 |
GO:0014823 |
response to activity(GO:0014823) |
| 0.0 |
0.3 |
GO:0043982 |
histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 |
0.2 |
GO:0006907 |
pinocytosis(GO:0006907) |
| 0.0 |
0.2 |
GO:0008088 |
axo-dendritic transport(GO:0008088) |
| 0.0 |
0.4 |
GO:0048387 |
negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 |
0.5 |
GO:0018214 |
peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 |
0.1 |
GO:0043012 |
regulation of fusion of sperm to egg plasma membrane(GO:0043012) |
| 0.0 |
0.3 |
GO:2001021 |
negative regulation of response to DNA damage stimulus(GO:2001021) |
| 0.0 |
0.0 |
GO:0002566 |
somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 |
0.2 |
GO:0015827 |
tryptophan transport(GO:0015827) leucine import(GO:0060356) |
| 0.0 |
0.1 |
GO:0071243 |
cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 |
1.5 |
GO:0015991 |
energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 |
0.2 |
GO:0000087 |
mitotic M phase(GO:0000087) |
| 0.0 |
1.1 |
GO:0072595 |
maintenance of protein location in nucleus(GO:0051457) maintenance of protein localization in organelle(GO:0072595) |
| 0.0 |
0.2 |
GO:0021854 |
hypothalamus development(GO:0021854) |
| 0.0 |
3.5 |
GO:0006977 |
DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) signal transduction involved in cell cycle checkpoint(GO:0072395) signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in mitotic cell cycle checkpoint(GO:0072413) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in mitotic G1 DNA damage checkpoint(GO:0072431) intracellular signal transduction involved in G1 DNA damage checkpoint(GO:1902400) signal transduction involved in mitotic DNA damage checkpoint(GO:1902402) signal transduction involved in mitotic DNA integrity checkpoint(GO:1902403) |
| 0.0 |
0.5 |
GO:0046697 |
decidualization(GO:0046697) |
| 0.0 |
0.9 |
GO:0018107 |
peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 |
0.4 |
GO:0042249 |
establishment of planar polarity of embryonic epithelium(GO:0042249) |
| 0.0 |
1.3 |
GO:0007026 |
negative regulation of microtubule depolymerization(GO:0007026) regulation of microtubule depolymerization(GO:0031114) negative regulation of protein depolymerization(GO:1901880) |
| 0.0 |
0.2 |
GO:0010993 |
ubiquitin homeostasis(GO:0010992) regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.0 |
0.1 |
GO:0018410 |
C-terminal protein amino acid modification(GO:0018410) |
| 0.0 |
0.2 |
GO:0045727 |
positive regulation of cellular amide metabolic process(GO:0034250) positive regulation of translation(GO:0045727) |
| 0.0 |
0.2 |
GO:0017183 |
peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 |
0.1 |
GO:0021506 |
anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.0 |
0.2 |
GO:0032375 |
negative regulation of sterol transport(GO:0032372) negative regulation of cholesterol transport(GO:0032375) |
| 0.0 |
0.1 |
GO:0034770 |
histone H4-K20 methylation(GO:0034770) |
| 0.0 |
0.5 |
GO:0043508 |
negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 |
0.1 |
GO:0000244 |
spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 |
0.4 |
GO:0006884 |
cell volume homeostasis(GO:0006884) |
| 0.0 |
0.5 |
GO:0032456 |
endocytic recycling(GO:0032456) |
| 0.0 |
0.1 |
GO:0007091 |
metaphase/anaphase transition of mitotic cell cycle(GO:0007091) regulation of mitotic sister chromatid separation(GO:0010965) regulation of mitotic sister chromatid segregation(GO:0033047) metaphase/anaphase transition of cell cycle(GO:0044784) chromosome separation(GO:0051304) mitotic sister chromatid separation(GO:0051306) |
| 0.0 |
0.0 |
GO:0045908 |
negative regulation of vasodilation(GO:0045908) |
| 0.0 |
2.2 |
GO:0006665 |
sphingolipid metabolic process(GO:0006665) |
| 0.0 |
0.4 |
GO:0048488 |
synaptic vesicle recycling(GO:0036465) synaptic vesicle endocytosis(GO:0048488) clathrin-mediated endocytosis(GO:0072583) |
| 0.0 |
0.3 |
GO:0045652 |
regulation of megakaryocyte differentiation(GO:0045652) regulation of hematopoietic progenitor cell differentiation(GO:1901532) |
| 0.0 |
0.2 |
GO:0090231 |
regulation of spindle checkpoint(GO:0090231) |
| 0.0 |
0.2 |
GO:0046628 |
positive regulation of insulin receptor signaling pathway(GO:0046628) positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 |
0.7 |
GO:0018208 |
peptidyl-proline modification(GO:0018208) |
| 0.0 |
0.4 |
GO:0006103 |
2-oxoglutarate metabolic process(GO:0006103) |
| 0.0 |
0.2 |
GO:0048024 |
regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.0 |
2.4 |
GO:0042035 |
regulation of cytokine biosynthetic process(GO:0042035) |
| 0.0 |
0.0 |
GO:0060136 |
embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 |
0.6 |
GO:0043968 |
histone H2A acetylation(GO:0043968) |
| 0.0 |
0.1 |
GO:0043583 |
ear development(GO:0043583) |
| 0.0 |
0.1 |
GO:0021527 |
spinal cord association neuron differentiation(GO:0021527) |
| 0.0 |
0.1 |
GO:0043547 |
positive regulation of GTPase activity(GO:0043547) |
| 0.0 |
0.1 |
GO:0034337 |
RNA folding(GO:0034337) DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) regulation of DNA methylation(GO:0044030) |
| 0.0 |
0.5 |
GO:0000768 |
syncytium formation by plasma membrane fusion(GO:0000768) myoblast fusion(GO:0007520) |
| 0.0 |
0.2 |
GO:0007584 |
response to nutrient(GO:0007584) |
| 0.0 |
0.3 |
GO:0060393 |
pathway-restricted SMAD protein phosphorylation(GO:0060389) regulation of pathway-restricted SMAD protein phosphorylation(GO:0060393) |
| 0.0 |
0.6 |
GO:0045446 |
endothelial cell differentiation(GO:0045446) |
| 0.0 |
0.3 |
GO:0015937 |
coenzyme A biosynthetic process(GO:0015937) |
| 0.0 |
0.1 |
GO:0010831 |
positive regulation of myotube differentiation(GO:0010831) |
| 0.0 |
0.2 |
GO:0098930 |
axonal transport(GO:0098930) |
| 0.0 |
0.3 |
GO:0006743 |
ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) |
| 0.0 |
0.1 |
GO:0045721 |
negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 |
0.1 |
GO:0015693 |
magnesium ion transport(GO:0015693) |
| 0.0 |
0.1 |
GO:0046952 |
cellular ketone body metabolic process(GO:0046950) ketone body catabolic process(GO:0046952) |
| 0.0 |
0.7 |
GO:0010332 |
response to gamma radiation(GO:0010332) |
| 0.0 |
1.5 |
GO:0006939 |
smooth muscle contraction(GO:0006939) |
| 0.0 |
0.1 |
GO:0006423 |
cysteinyl-tRNA aminoacylation(GO:0006423) |
| 0.0 |
2.2 |
GO:0018279 |
peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 |
0.1 |
GO:0010595 |
positive regulation of endothelial cell migration(GO:0010595) |
| 0.0 |
0.6 |
GO:0045995 |
regulation of embryonic development(GO:0045995) |
| 0.0 |
0.3 |
GO:0016226 |
iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 |
0.1 |
GO:0035405 |
histone-threonine phosphorylation(GO:0035405) |
| 0.0 |
0.9 |
GO:0050817 |
blood coagulation(GO:0007596) hemostasis(GO:0007599) coagulation(GO:0050817) |
| 0.0 |
0.1 |
GO:0033539 |
fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 |
0.2 |
GO:0043545 |
Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 |
0.2 |
GO:0015939 |
pantothenate metabolic process(GO:0015939) |
| 0.0 |
0.8 |
GO:1901264 |
carbohydrate derivative transport(GO:1901264) |
| 0.0 |
0.3 |
GO:0034643 |
establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 |
0.2 |
GO:0033119 |
negative regulation of RNA splicing(GO:0033119) |
| 0.0 |
0.7 |
GO:0043534 |
blood vessel endothelial cell migration(GO:0043534) |
| 0.0 |
0.3 |
GO:0006379 |
mRNA cleavage(GO:0006379) |
| 0.0 |
0.3 |
GO:0007253 |
cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 |
0.6 |
GO:0034199 |
activation of protein kinase A activity(GO:0034199) |
| 0.0 |
0.3 |
GO:0006449 |
regulation of translational termination(GO:0006449) |
| 0.0 |
0.3 |
GO:0050704 |
regulation of interleukin-1 secretion(GO:0050704) positive regulation of interleukin-1 secretion(GO:0050716) |
| 0.0 |
3.8 |
GO:0006936 |
muscle contraction(GO:0006936) |
| 0.0 |
0.1 |
GO:0007413 |
axonal fasciculation(GO:0007413) |
| 0.0 |
0.3 |
GO:0031648 |
protein destabilization(GO:0031648) |
| 0.0 |
0.1 |
GO:0009249 |
protein lipoylation(GO:0009249) |
| 0.0 |
0.5 |
GO:0006084 |
acetyl-CoA metabolic process(GO:0006084) |
| 0.0 |
0.4 |
GO:0007033 |
vacuole organization(GO:0007033) |
| 0.0 |
0.2 |
GO:0006890 |
retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 |
0.1 |
GO:0050790 |
regulation of catalytic activity(GO:0050790) |
| 0.0 |
0.5 |
GO:0005980 |
glycogen catabolic process(GO:0005980) |
| 0.0 |
0.1 |
GO:0015811 |
sulfur amino acid transport(GO:0000101) L-cystine transport(GO:0015811) |
| 0.0 |
0.2 |
GO:1904030 |
negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 |
0.1 |
GO:0016557 |
peroxisome membrane biogenesis(GO:0016557) |
| 0.0 |
0.1 |
GO:0006104 |
succinyl-CoA metabolic process(GO:0006104) |
| 0.0 |
0.1 |
GO:0000185 |
activation of MAPKKK activity(GO:0000185) |
| 0.0 |
0.2 |
GO:0006465 |
signal peptide processing(GO:0006465) |
| 0.0 |
0.0 |
GO:0046984 |
regulation of hemoglobin biosynthetic process(GO:0046984) |
| 0.0 |
0.4 |
GO:0031929 |
TOR signaling(GO:0031929) |
| 0.0 |
0.4 |
GO:0030574 |
collagen catabolic process(GO:0030574) |
| 0.0 |
0.0 |
GO:0060689 |
cell differentiation involved in salivary gland development(GO:0060689) |
| 0.0 |
0.2 |
GO:0001895 |
retina homeostasis(GO:0001895) photoreceptor cell maintenance(GO:0045494) |
| 0.0 |
0.1 |
GO:0070306 |
lens fiber cell differentiation(GO:0070306) |
| 0.0 |
0.0 |
GO:0000338 |
protein deneddylation(GO:0000338) |
| 0.0 |
0.4 |
GO:0007030 |
Golgi organization(GO:0007030) |
| 0.0 |
0.1 |
GO:0045161 |
neuronal ion channel clustering(GO:0045161) |
| 0.0 |
0.1 |
GO:0007626 |
locomotory behavior(GO:0007626) |
| 0.0 |
0.1 |
GO:0002032 |
desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 |
0.0 |
GO:0007616 |
long-term memory(GO:0007616) synaptic vesicle docking(GO:0016081) |
| 0.0 |
0.2 |
GO:0045600 |
positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 |
1.2 |
GO:0007218 |
neuropeptide signaling pathway(GO:0007218) |
| 0.0 |
0.2 |
GO:0006613 |
cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 |
0.4 |
GO:0000080 |
mitotic G1 phase(GO:0000080) |
| 0.0 |
0.1 |
GO:0051602 |
response to electrical stimulus(GO:0051602) |
| 0.0 |
0.4 |
GO:0001843 |
neural tube closure(GO:0001843) tube closure(GO:0060606) |
| 0.0 |
0.1 |
GO:0031635 |
adenylate cyclase-inhibiting opioid receptor signaling pathway(GO:0031635) opioid receptor signaling pathway(GO:0038003) |
| 0.0 |
0.1 |
GO:0031032 |
actomyosin structure organization(GO:0031032) |
| 0.0 |
1.8 |
GO:0007156 |
homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 |
0.1 |
GO:0034723 |
DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 |
0.1 |
GO:0035235 |
ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 |
1.0 |
GO:0008277 |
regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 |
0.2 |
GO:0006309 |
DNA catabolic process, endonucleolytic(GO:0000737) apoptotic DNA fragmentation(GO:0006309) |
| 0.0 |
0.0 |
GO:0030157 |
pancreatic juice secretion(GO:0030157) |
| 0.0 |
0.1 |
GO:0001510 |
RNA methylation(GO:0001510) |
| 0.0 |
0.1 |
GO:0000097 |
sulfur amino acid biosynthetic process(GO:0000097) methionine biosynthetic process(GO:0009086) |
| 0.0 |
0.0 |
GO:0060969 |
negative regulation of gene silencing(GO:0060969) |
| 0.0 |
0.1 |
GO:0051775 |
response to redox state(GO:0051775) |
| 0.0 |
0.0 |
GO:0006707 |
cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 |
0.1 |
GO:0050687 |
negative regulation of defense response to virus(GO:0050687) |
| 0.0 |
0.1 |
GO:2000398 |
regulation of T cell differentiation in thymus(GO:0033081) regulation of thymocyte aggregation(GO:2000398) |
| 0.0 |
0.4 |
GO:0046324 |
regulation of glucose import(GO:0046324) |
| 0.0 |
0.9 |
GO:2000116 |
regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043281) regulation of cysteine-type endopeptidase activity(GO:2000116) |
| 0.0 |
0.4 |
GO:0034976 |
response to endoplasmic reticulum stress(GO:0034976) |
| 0.0 |
0.0 |
GO:0006434 |
seryl-tRNA aminoacylation(GO:0006434) |