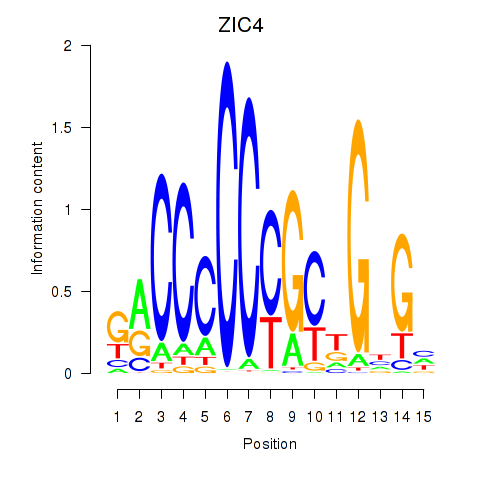
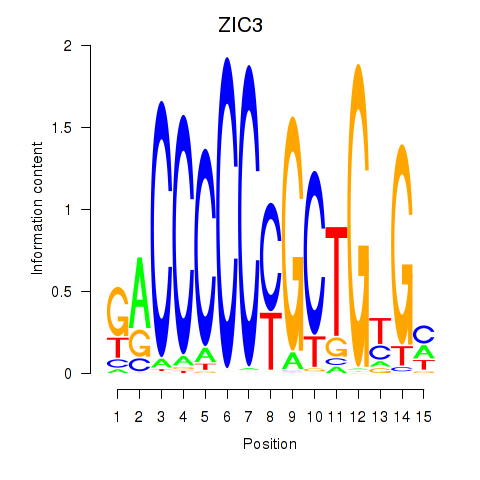
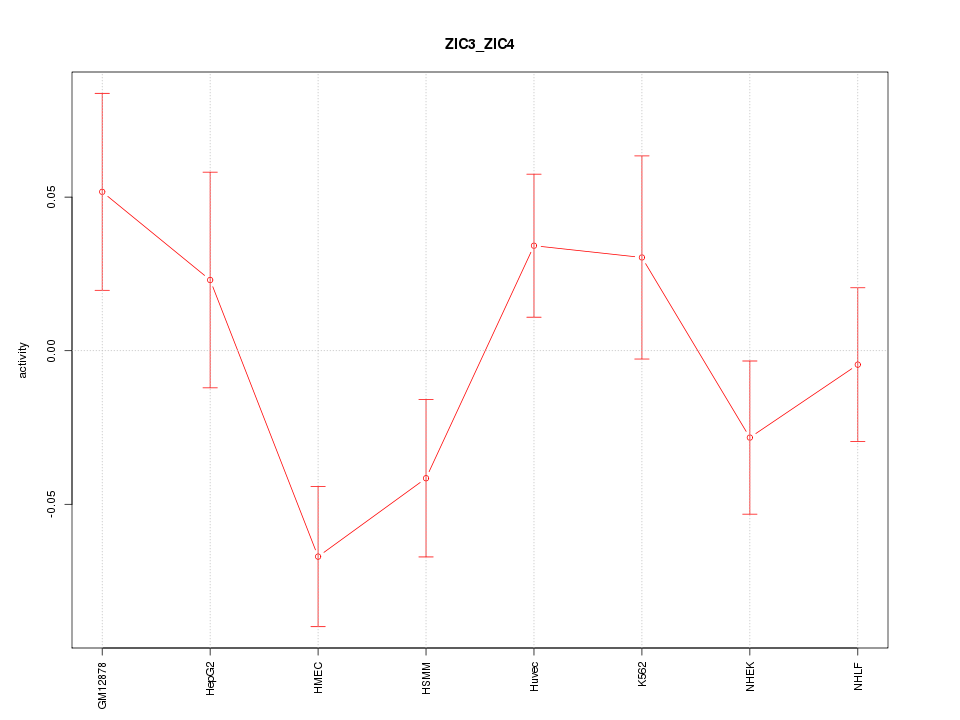
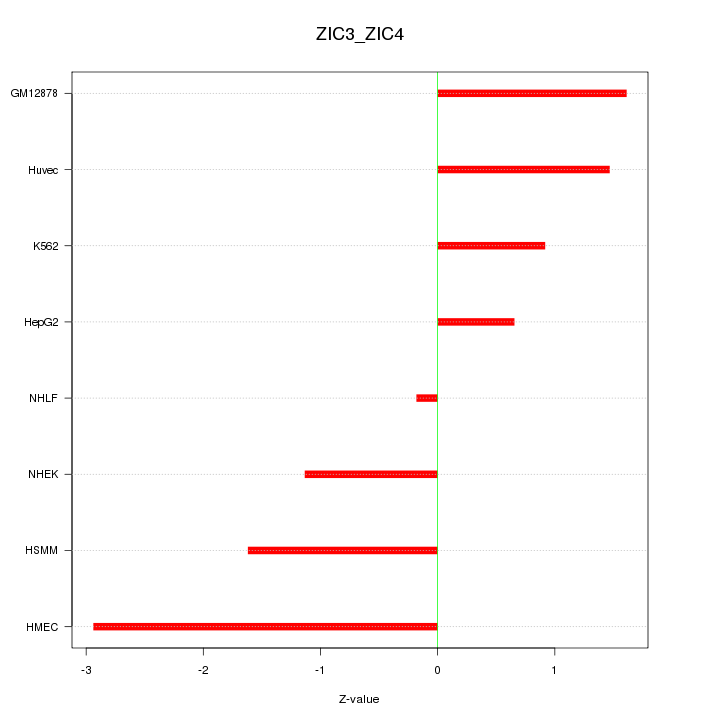
| 0.5 |
2.5 |
GO:0003093 |
regulation of glomerular filtration(GO:0003093) glomerular filtration(GO:0003094) renal filtration(GO:0097205) regulation of renal system process(GO:0098801) |
| 0.3 |
0.5 |
GO:0070672 |
response to interleukin-15(GO:0070672) |
| 0.2 |
0.7 |
GO:0021919 |
BMP signaling pathway involved in spinal cord dorsal/ventral patterning(GO:0021919) |
| 0.2 |
0.6 |
GO:0014887 |
muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.2 |
0.6 |
GO:0070671 |
response to interleukin-12(GO:0070671) |
| 0.2 |
2.1 |
GO:0090083 |
regulation of inclusion body assembly(GO:0090083) |
| 0.2 |
0.5 |
GO:0061030 |
epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.1 |
0.6 |
GO:0033602 |
negative regulation of gamma-aminobutyric acid secretion(GO:0014053) negative regulation of dopamine secretion(GO:0033602) |
| 0.1 |
0.4 |
GO:0060012 |
synaptic transmission, glycinergic(GO:0060012) |
| 0.1 |
0.4 |
GO:0070601 |
maintenance of DNA methylation(GO:0010216) centromeric sister chromatid cohesion(GO:0070601) regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.1 |
1.3 |
GO:0007130 |
synaptonemal complex assembly(GO:0007130) |
| 0.1 |
0.4 |
GO:2000644 |
low-density lipoprotein particle receptor catabolic process(GO:0032802) regulation of low-density lipoprotein particle receptor catabolic process(GO:0032803) negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) regulation of receptor catabolic process(GO:2000644) |
| 0.1 |
0.8 |
GO:0060272 |
embryonic skeletal joint morphogenesis(GO:0060272) embryonic skeletal joint development(GO:0072498) |
| 0.1 |
0.5 |
GO:0010157 |
response to chlorate(GO:0010157) |
| 0.1 |
0.4 |
GO:0031580 |
membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 |
0.3 |
GO:0009202 |
deoxyribonucleoside triphosphate biosynthetic process(GO:0009202) |
| 0.1 |
0.3 |
GO:0032328 |
alanine transport(GO:0032328) |
| 0.1 |
0.9 |
GO:0043306 |
positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of leukocyte degranulation(GO:0043302) positive regulation of mast cell degranulation(GO:0043306) |
| 0.1 |
0.3 |
GO:0006478 |
peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 |
0.3 |
GO:0001757 |
somite specification(GO:0001757) |
| 0.1 |
0.5 |
GO:0031665 |
negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.1 |
0.4 |
GO:0061028 |
establishment of endothelial barrier(GO:0061028) |
| 0.1 |
0.7 |
GO:0015870 |
acetylcholine transport(GO:0015870) acetate ester transport(GO:1901374) |
| 0.1 |
0.9 |
GO:0000076 |
DNA replication checkpoint(GO:0000076) |
| 0.1 |
0.6 |
GO:0034755 |
iron ion transmembrane transport(GO:0034755) |
| 0.1 |
0.3 |
GO:0032486 |
Rap protein signal transduction(GO:0032486) |
| 0.1 |
0.2 |
GO:0035608 |
protein deglutamylation(GO:0035608) |
| 0.1 |
1.1 |
GO:0050718 |
positive regulation of interleukin-1 beta secretion(GO:0050718) |
| 0.1 |
0.3 |
GO:0072106 |
regulation of ureteric bud formation(GO:0072106) positive regulation of ureteric bud formation(GO:0072107) |
| 0.1 |
0.2 |
GO:0010586 |
miRNA metabolic process(GO:0010586) miRNA catabolic process(GO:0010587) |
| 0.1 |
0.5 |
GO:0009820 |
alkaloid metabolic process(GO:0009820) |
| 0.1 |
0.3 |
GO:0006549 |
isoleucine metabolic process(GO:0006549) |
| 0.1 |
0.2 |
GO:0006669 |
sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.1 |
0.2 |
GO:0032927 |
positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.1 |
0.2 |
GO:0051823 |
regulation of synapse structural plasticity(GO:0051823) |
| 0.1 |
0.9 |
GO:0007035 |
vacuolar acidification(GO:0007035) |
| 0.1 |
0.4 |
GO:0032460 |
negative regulation of protein oligomerization(GO:0032460) negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 |
0.2 |
GO:0006384 |
transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 |
0.3 |
GO:0006857 |
oligopeptide transport(GO:0006857) |
| 0.1 |
0.4 |
GO:0070779 |
D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.1 |
0.2 |
GO:0051570 |
regulation of histone H3-K9 methylation(GO:0051570) |
| 0.1 |
0.4 |
GO:0021830 |
interneuron migration from the subpallium to the cortex(GO:0021830) cerebral cortex GABAergic interneuron migration(GO:0021853) cerebral cortex GABAergic interneuron development(GO:0021894) interneuron migration(GO:1904936) |
| 0.1 |
0.2 |
GO:0010792 |
DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) double-strand break repair via single-strand annealing(GO:0045002) |
| 0.1 |
0.2 |
GO:0015854 |
guanine transport(GO:0015854) hypoxanthine transport(GO:0035344) thymine transport(GO:0035364) |
| 0.1 |
0.2 |
GO:0019605 |
butyrate metabolic process(GO:0019605) |
| 0.1 |
0.3 |
GO:0007224 |
smoothened signaling pathway(GO:0007224) |
| 0.1 |
0.4 |
GO:0033326 |
cerebrospinal fluid secretion(GO:0033326) |
| 0.1 |
0.3 |
GO:0045943 |
positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 |
0.3 |
GO:0070192 |
synapsis(GO:0007129) chromosome organization involved in meiotic cell cycle(GO:0070192) |
| 0.0 |
0.5 |
GO:0015937 |
coenzyme A biosynthetic process(GO:0015937) |
| 0.0 |
0.6 |
GO:0032411 |
positive regulation of transporter activity(GO:0032411) |
| 0.0 |
0.2 |
GO:0045199 |
maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 |
0.5 |
GO:0032876 |
regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 |
0.2 |
GO:0046607 |
positive regulation of centrosome cycle(GO:0046607) |
| 0.0 |
0.1 |
GO:0033590 |
response to cobalamin(GO:0033590) |
| 0.0 |
2.2 |
GO:0060041 |
retina development in camera-type eye(GO:0060041) |
| 0.0 |
0.1 |
GO:0002514 |
B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) negative regulation of B cell mediated immunity(GO:0002713) negative regulation of immunoglobulin mediated immune response(GO:0002890) left/right pattern formation(GO:0060972) |
| 0.0 |
0.1 |
GO:0000447 |
endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) cleavage involved in rRNA processing(GO:0000469) endonucleolytic cleavage involved in rRNA processing(GO:0000478) endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 |
0.2 |
GO:0070935 |
positive regulation of interleukin-23 production(GO:0032747) 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 |
0.3 |
GO:0021675 |
nerve development(GO:0021675) |
| 0.0 |
0.2 |
GO:0090050 |
positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.0 |
0.2 |
GO:0051001 |
negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.0 |
0.2 |
GO:0051567 |
histone H3-K9 methylation(GO:0051567) histone H3-K9 modification(GO:0061647) |
| 0.0 |
0.1 |
GO:0045715 |
negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) negative regulation of lipoprotein metabolic process(GO:0050748) |
| 0.0 |
0.3 |
GO:0006020 |
inositol metabolic process(GO:0006020) |
| 0.0 |
0.1 |
GO:0048496 |
maintenance of organ identity(GO:0048496) |
| 0.0 |
0.4 |
GO:0018214 |
peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 |
0.1 |
GO:2000078 |
regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) regulation of type B pancreatic cell development(GO:2000074) positive regulation of type B pancreatic cell development(GO:2000078) |
| 0.0 |
0.4 |
GO:0051051 |
negative regulation of transport(GO:0051051) |
| 0.0 |
0.6 |
GO:0000303 |
response to superoxide(GO:0000303) |
| 0.0 |
0.4 |
GO:0009113 |
purine nucleobase biosynthetic process(GO:0009113) |
| 0.0 |
0.2 |
GO:0016080 |
synaptic vesicle targeting(GO:0016080) |
| 0.0 |
0.1 |
GO:0072284 |
visceral serous pericardium development(GO:0061032) S-shaped body morphogenesis(GO:0072050) metanephric S-shaped body morphogenesis(GO:0072284) |
| 0.0 |
0.1 |
GO:0007288 |
sperm axoneme assembly(GO:0007288) |
| 0.0 |
0.3 |
GO:0033363 |
secretory granule organization(GO:0033363) |
| 0.0 |
2.3 |
GO:0007605 |
sensory perception of sound(GO:0007605) |
| 0.0 |
0.1 |
GO:0055098 |
response to low-density lipoprotein particle(GO:0055098) regulation of cholesterol transporter activity(GO:0060694) |
| 0.0 |
0.1 |
GO:0071474 |
cellular water homeostasis(GO:0009992) cellular hyperosmotic response(GO:0071474) |
| 0.0 |
0.2 |
GO:0043488 |
regulation of RNA stability(GO:0043487) regulation of mRNA stability(GO:0043488) |
| 0.0 |
0.5 |
GO:0015701 |
bicarbonate transport(GO:0015701) |
| 0.0 |
0.2 |
GO:0006044 |
N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 |
0.0 |
GO:0060596 |
mammary placode formation(GO:0060596) |
| 0.0 |
0.3 |
GO:0007020 |
microtubule nucleation(GO:0007020) |
| 0.0 |
0.0 |
GO:0051066 |
dihydrobiopterin metabolic process(GO:0051066) |
| 0.0 |
0.1 |
GO:0007195 |
adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 |
0.1 |
GO:0019255 |
glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 |
0.4 |
GO:0051923 |
sulfation(GO:0051923) |
| 0.0 |
0.1 |
GO:0071894 |
histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.0 |
0.2 |
GO:0016601 |
Rac protein signal transduction(GO:0016601) |
| 0.0 |
0.2 |
GO:0006002 |
fructose 6-phosphate metabolic process(GO:0006002) |
| 0.0 |
0.1 |
GO:0046668 |
regulation of retinal cell programmed cell death(GO:0046668) |
| 0.0 |
0.3 |
GO:1903078 |
positive regulation of establishment of protein localization to plasma membrane(GO:0090004) positive regulation of protein localization to plasma membrane(GO:1903078) positive regulation of protein localization to cell periphery(GO:1904377) |
| 0.0 |
0.1 |
GO:0006102 |
isocitrate metabolic process(GO:0006102) |
| 0.0 |
0.4 |
GO:0006493 |
protein O-linked glycosylation(GO:0006493) |
| 0.0 |
0.1 |
GO:0060662 |
tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 |
0.1 |
GO:0015791 |
polyol transport(GO:0015791) |
| 0.0 |
0.1 |
GO:0043084 |
penile erection(GO:0043084) |
| 0.0 |
0.1 |
GO:0001885 |
endothelial cell development(GO:0001885) |
| 0.0 |
0.2 |
GO:0045540 |
regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.0 |
0.1 |
GO:0051592 |
response to calcium ion(GO:0051592) |
| 0.0 |
0.1 |
GO:0060423 |
foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) positive regulation of branching involved in lung morphogenesis(GO:0061047) iris morphogenesis(GO:0061072) cornea development in camera-type eye(GO:0061303) |
| 0.0 |
0.1 |
GO:0001706 |
endoderm formation(GO:0001706) |
| 0.0 |
1.4 |
GO:0031295 |
lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 |
0.5 |
GO:0006672 |
ceramide metabolic process(GO:0006672) |
| 0.0 |
0.3 |
GO:0032608 |
interferon-beta production(GO:0032608) regulation of interferon-beta production(GO:0032648) |
| 0.0 |
0.1 |
GO:0006927 |
obsolete transformed cell apoptotic process(GO:0006927) |
| 0.0 |
0.1 |
GO:0015851 |
nucleobase transport(GO:0015851) L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 |
4.4 |
GO:0051056 |
regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 |
0.9 |
GO:0008360 |
regulation of cell shape(GO:0008360) |
| 0.0 |
0.1 |
GO:0035058 |
positive regulation of neuroblast proliferation(GO:0002052) nonmotile primary cilium assembly(GO:0035058) |
| 0.0 |
0.6 |
GO:0071377 |
cellular response to glucagon stimulus(GO:0071377) |