Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
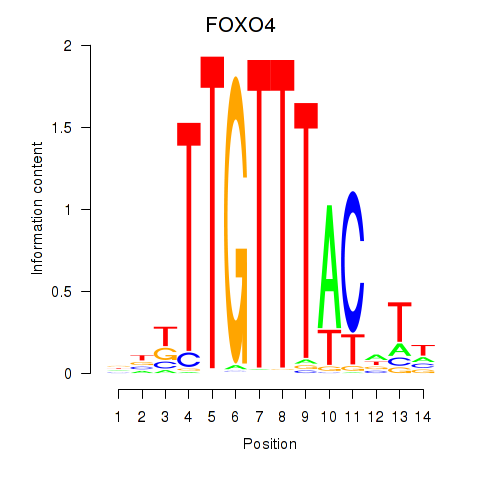
Results for FOXO4
Z-value: 1.73
Transcription factors associated with FOXO4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXO4
|
ENSG00000184481.12 | FOXO4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXO4 | hg19_v2_chrX_+_70316005_70316047 | -0.04 | 8.8e-01 | Click! |
Activity profile of FOXO4 motif
Sorted Z-values of FOXO4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXO4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_134551583 | 7.17 |
ENST00000435928.1 |
CALD1 |
caldesmon 1 |
| chrX_-_38080077 | 4.04 |
ENST00000378533.3 ENST00000544439.1 ENST00000432886.2 ENST00000538295.1 |
SRPX |
sushi-repeat containing protein, X-linked |
| chr7_-_95064264 | 3.72 |
ENST00000536183.1 ENST00000433091.2 ENST00000222572.3 |
PON2 |
paraoxonase 2 |
| chr3_+_159570722 | 3.35 |
ENST00000482804.1 |
SCHIP1 |
schwannomin interacting protein 1 |
| chr14_+_94577074 | 2.91 |
ENST00000444961.1 ENST00000448882.1 ENST00000557098.1 ENST00000554800.1 ENST00000556544.1 ENST00000298902.5 ENST00000555819.1 ENST00000557634.1 ENST00000555744.1 |
IFI27 |
interferon, alpha-inducible protein 27 |
| chr8_+_31497271 | 2.71 |
ENST00000520407.1 |
NRG1 |
neuregulin 1 |
| chr8_-_13134045 | 2.71 |
ENST00000512044.2 |
DLC1 |
deleted in liver cancer 1 |
| chr1_-_227505289 | 2.66 |
ENST00000366765.3 |
CDC42BPA |
CDC42 binding protein kinase alpha (DMPK-like) |
| chr11_+_844406 | 2.52 |
ENST00000397404.1 |
TSPAN4 |
tetraspanin 4 |
| chr9_+_706842 | 2.50 |
ENST00000382293.3 |
KANK1 |
KN motif and ankyrin repeat domains 1 |
| chr18_+_47088401 | 2.50 |
ENST00000261292.4 ENST00000427224.2 ENST00000580036.1 |
LIPG |
lipase, endothelial |
| chr12_+_96588143 | 2.42 |
ENST00000228741.3 ENST00000547249.1 |
ELK3 |
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr8_-_141931287 | 2.34 |
ENST00000517887.1 |
PTK2 |
protein tyrosine kinase 2 |
| chr12_-_10251576 | 2.33 |
ENST00000315330.4 |
CLEC1A |
C-type lectin domain family 1, member A |
| chr8_-_6420930 | 2.32 |
ENST00000325203.5 |
ANGPT2 |
angiopoietin 2 |
| chr8_-_6420777 | 2.32 |
ENST00000415216.1 |
ANGPT2 |
angiopoietin 2 |
| chr11_+_844067 | 2.28 |
ENST00000397406.1 ENST00000409543.2 ENST00000525201.1 |
TSPAN4 |
tetraspanin 4 |
| chr12_-_10251603 | 2.27 |
ENST00000457018.2 |
CLEC1A |
C-type lectin domain family 1, member A |
| chr12_-_71031185 | 2.18 |
ENST00000548122.1 ENST00000551525.1 ENST00000550358.1 |
PTPRB |
protein tyrosine phosphatase, receptor type, B |
| chr10_+_123923105 | 2.17 |
ENST00000368999.1 |
TACC2 |
transforming, acidic coiled-coil containing protein 2 |
| chr9_+_27109133 | 2.15 |
ENST00000519097.1 ENST00000380036.4 |
TEK |
TEK tyrosine kinase, endothelial |
| chr6_-_105627735 | 2.13 |
ENST00000254765.3 |
POPDC3 |
popeye domain containing 3 |
| chr2_-_192711968 | 2.11 |
ENST00000304141.4 |
SDPR |
serum deprivation response |
| chr12_-_10251539 | 2.05 |
ENST00000420265.2 |
CLEC1A |
C-type lectin domain family 1, member A |
| chr6_+_123110465 | 2.00 |
ENST00000539041.1 |
SMPDL3A |
sphingomyelin phosphodiesterase, acid-like 3A |
| chr4_-_70626430 | 1.94 |
ENST00000310613.3 |
SULT1B1 |
sulfotransferase family, cytosolic, 1B, member 1 |
| chr4_-_186696425 | 1.83 |
ENST00000430503.1 ENST00000319454.6 ENST00000450341.1 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr1_+_145524891 | 1.63 |
ENST00000369304.3 |
ITGA10 |
integrin, alpha 10 |
| chr10_+_123922941 | 1.63 |
ENST00000360561.3 |
TACC2 |
transforming, acidic coiled-coil containing protein 2 |
| chr4_-_70626314 | 1.53 |
ENST00000510821.1 |
SULT1B1 |
sulfotransferase family, cytosolic, 1B, member 1 |
| chr1_-_227505826 | 1.52 |
ENST00000334218.5 ENST00000366766.2 ENST00000366764.2 |
CDC42BPA |
CDC42 binding protein kinase alpha (DMPK-like) |
| chr8_-_6420565 | 1.50 |
ENST00000338312.6 |
ANGPT2 |
angiopoietin 2 |
| chr2_-_165424973 | 1.49 |
ENST00000543549.1 |
GRB14 |
growth factor receptor-bound protein 14 |
| chr4_-_90757364 | 1.47 |
ENST00000508895.1 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
| chr12_+_53491220 | 1.43 |
ENST00000548547.1 ENST00000301464.3 |
IGFBP6 |
insulin-like growth factor binding protein 6 |
| chr2_+_109204909 | 1.42 |
ENST00000393310.1 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
| chr11_-_89224299 | 1.40 |
ENST00000343727.5 ENST00000531342.1 ENST00000375979.3 |
NOX4 |
NADPH oxidase 4 |
| chr4_-_159080806 | 1.39 |
ENST00000590648.1 |
FAM198B |
family with sequence similarity 198, member B |
| chr4_-_186697044 | 1.38 |
ENST00000437304.2 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr13_-_99630233 | 1.38 |
ENST00000376460.1 ENST00000442173.1 |
DOCK9 |
dedicator of cytokinesis 9 |
| chr21_+_37507210 | 1.38 |
ENST00000290354.5 |
CBR3 |
carbonyl reductase 3 |
| chr6_-_46889694 | 1.37 |
ENST00000283296.7 ENST00000362015.4 ENST00000456426.2 |
GPR116 |
G protein-coupled receptor 116 |
| chr10_+_123923205 | 1.36 |
ENST00000369004.3 ENST00000260733.3 |
TACC2 |
transforming, acidic coiled-coil containing protein 2 |
| chr11_-_89224139 | 1.34 |
ENST00000413594.2 |
NOX4 |
NADPH oxidase 4 |
| chr4_+_146403912 | 1.29 |
ENST00000507367.1 ENST00000394092.2 ENST00000515385.1 |
SMAD1 |
SMAD family member 1 |
| chr11_-_89223883 | 1.27 |
ENST00000528341.1 |
NOX4 |
NADPH oxidase 4 |
| chr18_-_21852143 | 1.26 |
ENST00000399443.3 |
OSBPL1A |
oxysterol binding protein-like 1A |
| chr4_+_124320665 | 1.18 |
ENST00000394339.2 |
SPRY1 |
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
| chr13_+_73629107 | 1.18 |
ENST00000539231.1 |
KLF5 |
Kruppel-like factor 5 (intestinal) |
| chr1_-_57045228 | 1.16 |
ENST00000371250.3 |
PPAP2B |
phosphatidic acid phosphatase type 2B |
| chr11_-_85780853 | 1.15 |
ENST00000531930.1 ENST00000528398.1 |
PICALM |
phosphatidylinositol binding clathrin assembly protein |
| chr8_-_6420759 | 1.15 |
ENST00000523120.1 |
ANGPT2 |
angiopoietin 2 |
| chr20_+_44035200 | 1.13 |
ENST00000372717.1 ENST00000360981.4 |
DBNDD2 |
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr9_-_13175823 | 1.12 |
ENST00000545857.1 |
MPDZ |
multiple PDZ domain protein |
| chr11_+_77532233 | 1.12 |
ENST00000525409.1 |
AAMDC |
adipogenesis associated, Mth938 domain containing |
| chr5_-_125930929 | 1.11 |
ENST00000553117.1 ENST00000447989.2 ENST00000409134.3 |
ALDH7A1 |
aldehyde dehydrogenase 7 family, member A1 |
| chr1_-_168106536 | 1.10 |
ENST00000537209.1 ENST00000361697.2 ENST00000546300.1 ENST00000367835.1 |
GPR161 |
G protein-coupled receptor 161 |
| chr4_-_90756769 | 1.10 |
ENST00000345009.4 ENST00000505199.1 ENST00000502987.1 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
| chr20_+_44035847 | 1.10 |
ENST00000372712.2 |
DBNDD2 |
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr9_+_27109392 | 1.06 |
ENST00000406359.4 |
TEK |
TEK tyrosine kinase, endothelial |
| chr8_-_134309823 | 1.05 |
ENST00000414097.2 |
NDRG1 |
N-myc downstream regulated 1 |
| chr12_-_71031220 | 1.03 |
ENST00000334414.6 |
PTPRB |
protein tyrosine phosphatase, receptor type, B |
| chr12_+_96588279 | 1.02 |
ENST00000552142.1 |
ELK3 |
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr4_-_111119804 | 0.98 |
ENST00000394607.3 ENST00000302274.3 |
ELOVL6 |
ELOVL fatty acid elongase 6 |
| chr7_+_12250886 | 0.97 |
ENST00000444443.1 ENST00000396667.3 |
TMEM106B |
transmembrane protein 106B |
| chr2_+_109204743 | 0.97 |
ENST00000332345.6 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
| chr3_-_112127981 | 0.95 |
ENST00000486726.2 |
RP11-231E6.1 |
RP11-231E6.1 |
| chr18_-_53303123 | 0.91 |
ENST00000569357.1 ENST00000565124.1 ENST00000398339.1 |
TCF4 |
transcription factor 4 |
| chr8_-_134309335 | 0.90 |
ENST00000522890.1 ENST00000323851.7 ENST00000518176.1 ENST00000354944.5 ENST00000537882.1 ENST00000522476.1 ENST00000518066.1 ENST00000521544.1 ENST00000518480.1 ENST00000523892.1 |
NDRG1 |
N-myc downstream regulated 1 |
| chr3_+_173302222 | 0.88 |
ENST00000361589.4 |
NLGN1 |
neuroligin 1 |
| chr16_+_28858004 | 0.85 |
ENST00000322610.8 |
SH2B1 |
SH2B adaptor protein 1 |
| chr2_-_219157250 | 0.84 |
ENST00000434015.2 ENST00000444183.1 ENST00000420341.1 ENST00000453281.1 ENST00000258412.3 ENST00000440422.1 |
TMBIM1 |
transmembrane BAX inhibitor motif containing 1 |
| chr4_-_139163491 | 0.84 |
ENST00000280612.5 |
SLC7A11 |
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
| chr11_+_77532155 | 0.83 |
ENST00000532481.1 ENST00000526415.1 ENST00000393427.2 ENST00000527134.1 ENST00000304716.8 |
AAMDC |
adipogenesis associated, Mth938 domain containing |
| chr20_+_45338126 | 0.83 |
ENST00000359271.2 |
SLC2A10 |
solute carrier family 2 (facilitated glucose transporter), member 10 |
| chr14_+_96968707 | 0.81 |
ENST00000216277.8 ENST00000557320.1 ENST00000557471.1 |
PAPOLA |
poly(A) polymerase alpha |
| chr3_+_138068051 | 0.80 |
ENST00000474559.1 |
MRAS |
muscle RAS oncogene homolog |
| chr5_+_79703823 | 0.80 |
ENST00000338008.5 ENST00000510158.1 ENST00000505560.1 |
ZFYVE16 |
zinc finger, FYVE domain containing 16 |
| chr15_-_55657428 | 0.79 |
ENST00000568543.1 |
CCPG1 |
cell cycle progression 1 |
| chr3_-_28390581 | 0.78 |
ENST00000479665.1 |
AZI2 |
5-azacytidine induced 2 |
| chr19_-_36523709 | 0.78 |
ENST00000592017.1 ENST00000360535.4 |
CLIP3 |
CAP-GLY domain containing linker protein 3 |
| chr8_+_70404996 | 0.77 |
ENST00000402687.4 ENST00000419716.3 |
SULF1 |
sulfatase 1 |
| chr13_-_36944307 | 0.76 |
ENST00000355182.4 |
SPG20 |
spastic paraplegia 20 (Troyer syndrome) |
| chr1_+_25664408 | 0.75 |
ENST00000374358.4 |
TMEM50A |
transmembrane protein 50A |
| chr8_-_62602327 | 0.74 |
ENST00000445642.3 ENST00000517847.2 ENST00000389204.4 ENST00000517661.1 ENST00000517903.1 ENST00000522603.1 ENST00000522349.1 ENST00000522835.1 ENST00000541428.1 ENST00000518306.1 |
ASPH |
aspartate beta-hydroxylase |
| chr7_-_115670792 | 0.73 |
ENST00000265440.7 ENST00000393485.1 |
TFEC |
transcription factor EC |
| chr7_+_55177416 | 0.72 |
ENST00000450046.1 ENST00000454757.2 |
EGFR |
epidermal growth factor receptor |
| chr7_+_106505696 | 0.72 |
ENST00000440650.2 ENST00000496166.1 ENST00000473541.1 |
PIK3CG |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr2_+_65283529 | 0.72 |
ENST00000546106.1 ENST00000537589.1 ENST00000260569.4 |
CEP68 |
centrosomal protein 68kDa |
| chr7_-_115670804 | 0.71 |
ENST00000320239.7 |
TFEC |
transcription factor EC |
| chr6_-_31651817 | 0.70 |
ENST00000375863.3 ENST00000375860.2 |
LY6G5C |
lymphocyte antigen 6 complex, locus G5C |
| chr13_+_32838801 | 0.69 |
ENST00000542859.1 |
FRY |
furry homolog (Drosophila) |
| chr6_+_32121789 | 0.68 |
ENST00000437001.2 ENST00000375137.2 |
PPT2 |
palmitoyl-protein thioesterase 2 |
| chr17_+_17876127 | 0.66 |
ENST00000582416.1 ENST00000313838.8 ENST00000411504.2 ENST00000581264.1 ENST00000399187.1 ENST00000479684.2 ENST00000584166.1 ENST00000585108.1 ENST00000399182.1 ENST00000579977.1 |
LRRC48 |
leucine rich repeat containing 48 |
| chr14_-_55878538 | 0.65 |
ENST00000247178.5 |
ATG14 |
autophagy related 14 |
| chr17_-_17875688 | 0.64 |
ENST00000379504.3 ENST00000318094.10 ENST00000540946.1 ENST00000542206.1 ENST00000395739.4 ENST00000581396.1 ENST00000535933.1 ENST00000579586.1 |
TOM1L2 |
target of myb1-like 2 (chicken) |
| chr1_-_43855479 | 0.64 |
ENST00000290663.6 ENST00000372457.4 |
MED8 |
mediator complex subunit 8 |
| chr17_+_17082842 | 0.62 |
ENST00000579361.1 |
MPRIP |
myosin phosphatase Rho interacting protein |
| chr6_-_31830655 | 0.62 |
ENST00000375631.4 |
NEU1 |
sialidase 1 (lysosomal sialidase) |
| chr6_-_167571817 | 0.61 |
ENST00000366834.1 |
GPR31 |
G protein-coupled receptor 31 |
| chr13_-_36920420 | 0.60 |
ENST00000438666.2 |
SPG20 |
spastic paraplegia 20 (Troyer syndrome) |
| chr2_+_233527443 | 0.60 |
ENST00000410095.1 |
EFHD1 |
EF-hand domain family, member D1 |
| chr2_+_87135076 | 0.59 |
ENST00000409776.2 |
RGPD1 |
RANBP2-like and GRIP domain containing 1 |
| chr1_-_43855444 | 0.59 |
ENST00000372455.4 |
MED8 |
mediator complex subunit 8 |
| chr2_+_65283506 | 0.58 |
ENST00000377990.2 |
CEP68 |
centrosomal protein 68kDa |
| chr6_+_32121908 | 0.58 |
ENST00000375143.2 ENST00000424499.1 |
PPT2 |
palmitoyl-protein thioesterase 2 |
| chr6_-_42016385 | 0.57 |
ENST00000502771.1 ENST00000508143.1 ENST00000514588.1 ENST00000510503.1 ENST00000415497.2 ENST00000372988.4 |
CCND3 |
cyclin D3 |
| chr2_+_66662510 | 0.56 |
ENST00000272369.9 ENST00000407092.2 |
MEIS1 |
Meis homeobox 1 |
| chr4_-_164534657 | 0.56 |
ENST00000339875.5 |
MARCH1 |
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr6_+_101846664 | 0.56 |
ENST00000421544.1 ENST00000413795.1 ENST00000369138.1 ENST00000358361.3 |
GRIK2 |
glutamate receptor, ionotropic, kainate 2 |
| chr8_+_120079478 | 0.55 |
ENST00000332843.2 |
COLEC10 |
collectin sub-family member 10 (C-type lectin) |
| chr3_-_185826855 | 0.55 |
ENST00000306376.5 |
ETV5 |
ets variant 5 |
| chr18_+_28898052 | 0.54 |
ENST00000257192.4 |
DSG1 |
desmoglein 1 |
| chr3_+_138067666 | 0.51 |
ENST00000475711.1 ENST00000464896.1 |
MRAS |
muscle RAS oncogene homolog |
| chr20_-_62582475 | 0.49 |
ENST00000369908.5 |
UCKL1 |
uridine-cytidine kinase 1-like 1 |
| chr1_-_241803649 | 0.49 |
ENST00000366554.2 |
OPN3 |
opsin 3 |
| chr20_-_43150601 | 0.48 |
ENST00000541235.1 ENST00000255175.1 ENST00000342374.4 |
SERINC3 |
serine incorporator 3 |
| chr1_-_241803679 | 0.47 |
ENST00000331838.5 |
OPN3 |
opsin 3 |
| chr4_-_102268628 | 0.46 |
ENST00000323055.6 ENST00000512215.1 ENST00000394854.3 |
PPP3CA |
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr16_+_6069072 | 0.46 |
ENST00000547605.1 ENST00000550418.1 ENST00000553186.1 |
RBFOX1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr3_+_101504200 | 0.46 |
ENST00000422132.1 |
NXPE3 |
neurexophilin and PC-esterase domain family, member 3 |
| chr7_+_106505912 | 0.46 |
ENST00000359195.3 |
PIK3CG |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr14_+_50234827 | 0.45 |
ENST00000554589.1 ENST00000557247.1 |
KLHDC2 |
kelch domain containing 2 |
| chr9_+_5510492 | 0.42 |
ENST00000397745.2 |
PDCD1LG2 |
programmed cell death 1 ligand 2 |
| chr12_-_12491608 | 0.41 |
ENST00000545735.1 |
MANSC1 |
MANSC domain containing 1 |
| chr2_-_88285309 | 0.41 |
ENST00000420840.2 |
RGPD2 |
RANBP2-like and GRIP domain containing 2 |
| chr1_+_43855560 | 0.39 |
ENST00000562955.1 |
SZT2 |
seizure threshold 2 homolog (mouse) |
| chr5_-_141338627 | 0.39 |
ENST00000231484.3 |
PCDH12 |
protocadherin 12 |
| chr2_+_175260451 | 0.38 |
ENST00000458563.1 ENST00000409673.3 ENST00000272732.6 ENST00000435964.1 |
SCRN3 |
secernin 3 |
| chr19_+_17378159 | 0.38 |
ENST00000598188.1 ENST00000359435.4 ENST00000599474.1 ENST00000599057.1 ENST00000601043.1 ENST00000447614.2 |
BABAM1 |
BRISC and BRCA1 A complex member 1 |
| chr19_+_30863271 | 0.38 |
ENST00000355537.3 |
ZNF536 |
zinc finger protein 536 |
| chr2_+_12857043 | 0.38 |
ENST00000381465.2 |
TRIB2 |
tribbles pseudokinase 2 |
| chr1_+_151739131 | 0.37 |
ENST00000400999.1 |
OAZ3 |
ornithine decarboxylase antizyme 3 |
| chr19_+_17378278 | 0.36 |
ENST00000596335.1 ENST00000601436.1 ENST00000595632.1 |
BABAM1 |
BRISC and BRCA1 A complex member 1 |
| chr19_+_859425 | 0.36 |
ENST00000327726.6 |
CFD |
complement factor D (adipsin) |
| chr2_-_71454185 | 0.35 |
ENST00000244221.8 |
PAIP2B |
poly(A) binding protein interacting protein 2B |
| chrX_-_20236970 | 0.35 |
ENST00000379548.4 |
RPS6KA3 |
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr18_-_53069419 | 0.33 |
ENST00000570177.2 |
TCF4 |
transcription factor 4 |
| chr19_-_58662139 | 0.33 |
ENST00000598312.1 |
ZNF329 |
zinc finger protein 329 |
| chr3_-_18466026 | 0.33 |
ENST00000417717.2 |
SATB1 |
SATB homeobox 1 |
| chr1_-_57431679 | 0.33 |
ENST00000371237.4 ENST00000535057.1 ENST00000543257.1 |
C8B |
complement component 8, beta polypeptide |
| chr2_+_201980827 | 0.33 |
ENST00000309955.3 ENST00000443227.1 ENST00000341222.6 ENST00000355558.4 ENST00000340870.5 ENST00000341582.6 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
| chr3_+_132316081 | 0.32 |
ENST00000249887.2 |
ACKR4 |
atypical chemokine receptor 4 |
| chr19_+_859654 | 0.32 |
ENST00000592860.1 |
CFD |
complement factor D (adipsin) |
| chr5_-_59064458 | 0.32 |
ENST00000502575.1 ENST00000507116.1 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
| chr2_-_101767715 | 0.31 |
ENST00000376840.4 ENST00000409318.1 |
TBC1D8 |
TBC1 domain family, member 8 (with GRAM domain) |
| chr16_+_6069586 | 0.31 |
ENST00000547372.1 |
RBFOX1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr4_+_88754069 | 0.30 |
ENST00000395102.4 ENST00000497649.2 |
MEPE |
matrix extracellular phosphoglycoprotein |
| chr8_+_19171128 | 0.30 |
ENST00000265807.3 |
SH2D4A |
SH2 domain containing 4A |
| chr1_-_153514241 | 0.30 |
ENST00000368718.1 ENST00000359215.1 |
S100A5 |
S100 calcium binding protein A5 |
| chr1_-_53608249 | 0.30 |
ENST00000371494.4 |
SLC1A7 |
solute carrier family 1 (glutamate transporter), member 7 |
| chr6_-_117747015 | 0.30 |
ENST00000368508.3 ENST00000368507.3 |
ROS1 |
c-ros oncogene 1 , receptor tyrosine kinase |
| chr17_+_72426891 | 0.30 |
ENST00000392627.1 |
GPRC5C |
G protein-coupled receptor, family C, group 5, member C |
| chr1_+_28879588 | 0.29 |
ENST00000373830.3 |
TRNAU1AP |
tRNA selenocysteine 1 associated protein 1 |
| chr9_+_5510558 | 0.29 |
ENST00000397747.3 |
PDCD1LG2 |
programmed cell death 1 ligand 2 |
| chr2_+_12857015 | 0.29 |
ENST00000155926.4 |
TRIB2 |
tribbles pseudokinase 2 |
| chr1_+_73771844 | 0.29 |
ENST00000440762.1 ENST00000444827.1 ENST00000415686.1 ENST00000411903.1 |
RP4-598G3.1 |
RP4-598G3.1 |
| chr10_+_99400443 | 0.29 |
ENST00000370631.3 |
PI4K2A |
phosphatidylinositol 4-kinase type 2 alpha |
| chr1_+_6845384 | 0.28 |
ENST00000303635.7 |
CAMTA1 |
calmodulin binding transcription activator 1 |
| chr1_+_86934526 | 0.28 |
ENST00000394711.1 |
CLCA1 |
chloride channel accessory 1 |
| chr3_+_48956249 | 0.28 |
ENST00000452882.1 ENST00000430423.1 ENST00000356401.4 ENST00000449376.1 ENST00000420814.1 ENST00000449729.1 ENST00000433170.1 |
ARIH2 |
ariadne RBR E3 ubiquitin protein ligase 2 |
| chr7_-_6098770 | 0.27 |
ENST00000536084.1 ENST00000446699.1 ENST00000199389.6 |
EIF2AK1 |
eukaryotic translation initiation factor 2-alpha kinase 1 |
| chr3_+_130569592 | 0.26 |
ENST00000533801.2 |
ATP2C1 |
ATPase, Ca++ transporting, type 2C, member 1 |
| chr14_+_96968802 | 0.26 |
ENST00000556619.1 ENST00000392990.2 |
PAPOLA |
poly(A) polymerase alpha |
| chr4_+_88754113 | 0.25 |
ENST00000560249.1 ENST00000540395.1 ENST00000511670.1 ENST00000361056.3 |
MEPE |
matrix extracellular phosphoglycoprotein |
| chr7_-_38394118 | 0.25 |
ENST00000390345.2 |
TRGV4 |
T cell receptor gamma variable 4 |
| chr11_-_114466471 | 0.25 |
ENST00000424261.2 |
NXPE4 |
neurexophilin and PC-esterase domain family, member 4 |
| chr9_+_134000948 | 0.25 |
ENST00000359428.5 ENST00000411637.2 ENST00000451030.1 |
NUP214 |
nucleoporin 214kDa |
| chrX_-_148676974 | 0.24 |
ENST00000524178.1 |
HSFX2 |
heat shock transcription factor family, X linked 2 |
| chr14_+_22985251 | 0.23 |
ENST00000390510.1 |
TRAJ27 |
T cell receptor alpha joining 27 |
| chrX_+_148855726 | 0.23 |
ENST00000370416.4 |
HSFX1 |
heat shock transcription factor family, X linked 1 |
| chr7_+_101460882 | 0.23 |
ENST00000292535.7 ENST00000549414.2 ENST00000550008.2 ENST00000546411.2 ENST00000556210.1 |
CUX1 |
cut-like homeobox 1 |
| chr19_-_40791302 | 0.23 |
ENST00000392038.2 ENST00000578123.1 |
AKT2 |
v-akt murine thymoma viral oncogene homolog 2 |
| chr13_-_41240717 | 0.23 |
ENST00000379561.5 |
FOXO1 |
forkhead box O1 |
| chr1_+_154193325 | 0.22 |
ENST00000428931.1 ENST00000441890.1 ENST00000271877.7 ENST00000412596.1 ENST00000368504.1 ENST00000437652.1 |
UBAP2L |
ubiquitin associated protein 2-like |
| chr16_-_28608424 | 0.22 |
ENST00000335715.4 |
SULT1A2 |
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2 |
| chr15_+_43985725 | 0.21 |
ENST00000413453.2 |
CKMT1A |
creatine kinase, mitochondrial 1A |
| chr5_-_133510456 | 0.21 |
ENST00000520417.1 |
SKP1 |
S-phase kinase-associated protein 1 |
| chr10_+_111985713 | 0.21 |
ENST00000239007.7 |
MXI1 |
MAX interactor 1, dimerization protein |
| chr5_+_140529630 | 0.21 |
ENST00000543635.1 |
PCDHB6 |
protocadherin beta 6 |
| chr17_+_44668035 | 0.21 |
ENST00000398238.4 ENST00000225282.8 |
NSF |
N-ethylmaleimide-sensitive factor |
| chr19_-_40732594 | 0.20 |
ENST00000430325.2 ENST00000433940.1 |
CNTD2 |
cyclin N-terminal domain containing 2 |
| chr11_-_10879572 | 0.20 |
ENST00000413761.2 |
ZBED5 |
zinc finger, BED-type containing 5 |
| chr16_+_14980632 | 0.20 |
ENST00000565655.1 |
NOMO1 |
NODAL modulator 1 |
| chr11_+_46638805 | 0.20 |
ENST00000434074.1 ENST00000312040.4 ENST00000451945.1 |
ATG13 |
autophagy related 13 |
| chr3_+_130569429 | 0.20 |
ENST00000505330.1 ENST00000504381.1 ENST00000507488.2 ENST00000393221.4 |
ATP2C1 |
ATPase, Ca++ transporting, type 2C, member 1 |
| chr5_+_162932554 | 0.19 |
ENST00000321757.6 ENST00000421814.2 ENST00000518095.1 |
MAT2B |
methionine adenosyltransferase II, beta |
| chr11_-_114466477 | 0.19 |
ENST00000375478.3 |
NXPE4 |
neurexophilin and PC-esterase domain family, member 4 |
| chr7_-_38389573 | 0.19 |
ENST00000390344.2 |
TRGV5 |
T cell receptor gamma variable 5 |
| chr1_+_24018269 | 0.18 |
ENST00000374550.3 |
RPL11 |
ribosomal protein L11 |
| chr10_-_118032697 | 0.18 |
ENST00000439649.3 |
GFRA1 |
GDNF family receptor alpha 1 |
| chr16_-_28857677 | 0.18 |
ENST00000313511.3 |
TUFM |
Tu translation elongation factor, mitochondrial |
| chr3_-_150920979 | 0.18 |
ENST00000309180.5 ENST00000480322.1 |
GPR171 |
G protein-coupled receptor 171 |
| chr6_-_87804815 | 0.17 |
ENST00000369582.2 |
CGA |
glycoprotein hormones, alpha polypeptide |
| chr4_-_70518941 | 0.17 |
ENST00000286604.4 ENST00000505512.1 ENST00000514019.1 |
UGT2A1 UGT2A1 |
UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus |
| chr9_-_15472730 | 0.17 |
ENST00000481862.1 |
PSIP1 |
PC4 and SFRS1 interacting protein 1 |
| chr15_-_58571445 | 0.17 |
ENST00000558231.1 |
ALDH1A2 |
aldehyde dehydrogenase 1 family, member A2 |
| chr10_-_118032979 | 0.17 |
ENST00000355422.6 |
GFRA1 |
GDNF family receptor alpha 1 |
| chr1_-_182360498 | 0.16 |
ENST00000417584.2 |
GLUL |
glutamate-ammonia ligase |
| chr4_+_74347400 | 0.16 |
ENST00000226355.3 |
AFM |
afamin |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 7.3 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 1.1 | 3.2 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.8 | 2.5 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.6 | 2.5 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.5 | 2.6 | GO:2000470 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) regulation of peroxidase activity(GO:2000468) positive regulation of peroxidase activity(GO:2000470) |
| 0.5 | 4.0 | GO:0001845 | phagolysosome assembly(GO:0001845) negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.4 | 1.2 | GO:0060940 | epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 0.4 | 1.2 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.4 | 2.7 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.3 | 1.4 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.3 | 0.9 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.3 | 1.4 | GO:0042374 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.3 | 2.3 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.2 | 0.7 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.2 | 0.7 | GO:0043006 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.2 | 0.8 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 0.2 | 2.7 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.2 | 2.0 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.2 | 2.2 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.2 | 1.2 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.2 | 1.0 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.2 | 3.7 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.2 | 0.8 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.2 | 1.3 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.2 | 0.8 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.2 | 0.5 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.1 | 3.7 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 1.1 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 1.0 | GO:0042426 | choline catabolic process(GO:0042426) |
| 0.1 | 0.7 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.1 | 1.2 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.1 | 0.7 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.1 | 4.0 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.1 | 0.6 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.1 | 1.1 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 | 0.4 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.1 | 0.3 | GO:0002276 | basophil activation involved in immune response(GO:0002276) |
| 0.1 | 1.1 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 | 0.3 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 0.1 | 0.7 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.1 | 0.5 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.1 | 0.5 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.1 | 0.3 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 0.6 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.1 | 2.9 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.1 | 0.2 | GO:1902617 | response to fluoride(GO:1902617) |
| 0.1 | 0.3 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.1 | 1.3 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 0.5 | GO:0009597 | detection of virus(GO:0009597) |
| 0.1 | 1.3 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.1 | 0.3 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 1.0 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 2.4 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.1 | 0.7 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.2 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 1.5 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.2 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 1.4 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.7 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 1.1 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 1.1 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 1.0 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.2 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.3 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 1.5 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.0 | 5.2 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 0.0 | 0.3 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.1 | GO:1900535 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.0 | 0.2 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.6 | GO:0060044 | negative regulation of cardiac muscle tissue growth(GO:0055022) negative regulation of cardiac muscle cell proliferation(GO:0060044) negative regulation of heart growth(GO:0061117) |
| 0.0 | 0.8 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.7 | GO:0032607 | interferon-alpha production(GO:0032607) |
| 0.0 | 0.1 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.2 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 1.3 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.4 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 1.7 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.3 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.8 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 1.1 | GO:0031440 | regulation of mRNA 3'-end processing(GO:0031440) |
| 0.0 | 0.2 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.0 | 1.0 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.6 | GO:0046626 | regulation of insulin receptor signaling pathway(GO:0046626) |
| 0.0 | 0.2 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.2 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.3 | GO:0043555 | regulation of translation in response to stress(GO:0043555) |
| 0.0 | 0.5 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.0 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.0 | 0.3 | GO:0060004 | reflex(GO:0060004) |
| 0.0 | 2.0 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.8 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.3 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.2 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.8 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.8 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.0 | 0.1 | GO:0097242 | beta-amyloid clearance(GO:0097242) |
| 0.0 | 0.0 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.0 | 0.2 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.2 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.6 | GO:0050918 | positive chemotaxis(GO:0050918) |
| 0.0 | 0.2 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.5 | 7.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.4 | 1.2 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.2 | 0.7 | GO:0097489 | multivesicular body, internal vesicle lumen(GO:0097489) |
| 0.2 | 4.0 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.2 | 0.7 | GO:1903349 | omegasome membrane(GO:1903349) |
| 0.2 | 1.2 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 0.7 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 0.7 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 2.6 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 0.2 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.1 | 2.7 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 9.7 | GO:0042641 | actomyosin(GO:0042641) |
| 0.1 | 0.3 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.1 | 0.5 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.4 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 0.2 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 0.9 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 4.2 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 1.1 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.3 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.2 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 4.2 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 1.2 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.2 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 4.3 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 3.3 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 1.9 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 2.2 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.3 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.0 | 0.7 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.6 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 11.6 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.3 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.3 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 1.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 1.4 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 1.8 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.1 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.0 | 0.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 1.4 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.8 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 1.1 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.7 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 1.1 | 3.4 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.5 | 2.6 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.5 | 1.4 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.4 | 4.0 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.3 | 1.6 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.3 | 2.5 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.3 | 2.7 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.3 | 7.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.2 | 2.0 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.2 | 0.8 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.2 | 0.8 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 1.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.2 | 0.8 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.2 | 1.0 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.2 | 3.7 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.2 | 1.3 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.2 | 0.6 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.1 | 0.7 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 2.3 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 1.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 8.8 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 3.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 3.2 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 0.6 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 1.2 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 1.0 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 1.2 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 2.6 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.3 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 1.1 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.1 | 0.3 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 0.6 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 0.3 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.1 | 0.5 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 2.7 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 2.0 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 0.3 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.9 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.8 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 2.5 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 1.3 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.3 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.5 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.5 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.8 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.5 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 1.3 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 3.5 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.2 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.5 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 1.2 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.5 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 1.1 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.6 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 2.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.7 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 0.7 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 5.1 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.0 | 0.3 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 3.2 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 6.4 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.1 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.1 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.2 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.4 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 3.6 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.6 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 1.2 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.2 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 14.4 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 3.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 1.1 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 2.8 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.6 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 3.0 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 1.2 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 6.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.7 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 2.4 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 3.4 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.3 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 1.5 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 1.0 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 5.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 2.0 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.2 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.3 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.9 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.3 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.4 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 12.0 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.2 | 0.7 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.1 | 3.7 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 2.7 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.1 | 6.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.0 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 2.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.9 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.1 | 1.1 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 1.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 1.9 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 1.6 | REACTOME G PROTEIN BETA GAMMA SIGNALLING | Genes involved in G-protein beta:gamma signalling |
| 0.0 | 1.1 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.8 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 1.2 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 1.2 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.7 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.6 | REACTOME ACTIVATION OF KAINATE RECEPTORS UPON GLUTAMATE BINDING | Genes involved in Activation of Kainate Receptors upon glutamate binding |
| 0.0 | 0.5 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 2.6 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 2.4 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 2.5 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.7 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.2 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.8 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.2 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 1.6 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.4 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.1 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.3 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.4 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.6 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.0 | 1.3 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.1 | REACTOME NEF MEDIATES DOWN MODULATION OF CELL SURFACE RECEPTORS BY RECRUITING THEM TO CLATHRIN ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 0.0 | 0.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |