Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
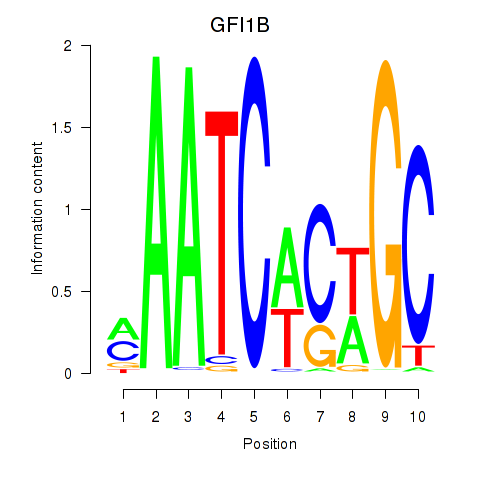
Results for GFI1B
Z-value: 2.16
Transcription factors associated with GFI1B
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GFI1B
|
ENSG00000165702.8 | GFI1B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GFI1B | hg19_v2_chr9_+_135854091_135854159 | -0.73 | 1.3e-03 | Click! |
Activity profile of GFI1B motif
Sorted Z-values of GFI1B motif
Network of associatons between targets according to the STRING database.
First level regulatory network of GFI1B
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_149688655 | 6.70 |
ENST00000461930.1 ENST00000423691.2 ENST00000490975.1 ENST00000461868.1 ENST00000452853.2 |
PFN2 |
profilin 2 |
| chr9_-_14180778 | 6.12 |
ENST00000380924.1 ENST00000543693.1 |
NFIB |
nuclear factor I/B |
| chr3_-_149688896 | 6.11 |
ENST00000239940.7 |
PFN2 |
profilin 2 |
| chr10_+_124221036 | 5.29 |
ENST00000368984.3 |
HTRA1 |
HtrA serine peptidase 1 |
| chr10_+_17272608 | 5.14 |
ENST00000421459.2 |
VIM |
vimentin |
| chr6_+_12290586 | 5.12 |
ENST00000379375.5 |
EDN1 |
endothelin 1 |
| chr14_-_53417732 | 4.85 |
ENST00000399304.3 ENST00000395631.2 ENST00000341590.3 ENST00000343279.4 |
FERMT2 |
fermitin family member 2 |
| chr7_+_134576317 | 4.22 |
ENST00000424922.1 ENST00000495522.1 |
CALD1 |
caldesmon 1 |
| chr1_-_68698222 | 3.96 |
ENST00000370976.3 ENST00000354777.2 ENST00000262348.4 ENST00000540432.1 |
WLS |
wntless Wnt ligand secretion mediator |
| chr7_+_134576151 | 3.71 |
ENST00000393118.2 |
CALD1 |
caldesmon 1 |
| chr7_+_79765071 | 3.69 |
ENST00000457358.2 |
GNAI1 |
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chr6_+_116601265 | 3.47 |
ENST00000452085.3 |
DSE |
dermatan sulfate epimerase |
| chr2_-_110371777 | 3.42 |
ENST00000397712.2 |
SEPT10 |
septin 10 |
| chr6_+_21593972 | 3.41 |
ENST00000244745.1 ENST00000543472.1 |
SOX4 |
SRY (sex determining region Y)-box 4 |
| chr2_-_110371720 | 3.35 |
ENST00000356688.4 |
SEPT10 |
septin 10 |
| chr11_+_32112431 | 3.34 |
ENST00000054950.3 |
RCN1 |
reticulocalbin 1, EF-hand calcium binding domain |
| chr2_-_110371412 | 3.21 |
ENST00000415095.1 ENST00000334001.6 ENST00000437928.1 ENST00000493445.1 ENST00000397714.2 ENST00000461295.1 |
SEPT10 |
septin 10 |
| chr1_-_94703118 | 3.20 |
ENST00000260526.6 ENST00000370217.3 |
ARHGAP29 |
Rho GTPase activating protein 29 |
| chr2_-_190044480 | 3.12 |
ENST00000374866.3 |
COL5A2 |
collagen, type V, alpha 2 |
| chr6_-_56707943 | 3.01 |
ENST00000370769.4 ENST00000421834.2 ENST00000312431.6 ENST00000361203.3 ENST00000523817.1 |
DST |
dystonin |
| chr1_+_101702417 | 2.89 |
ENST00000305352.6 |
S1PR1 |
sphingosine-1-phosphate receptor 1 |
| chr17_-_33759509 | 2.87 |
ENST00000304905.5 |
SLFN12 |
schlafen family member 12 |
| chr14_-_93214988 | 2.82 |
ENST00000557434.1 ENST00000393218.2 ENST00000334869.4 |
LGMN |
legumain |
| chr17_-_1395954 | 2.66 |
ENST00000359786.5 |
MYO1C |
myosin IC |
| chr14_-_93214915 | 2.65 |
ENST00000553918.1 ENST00000555699.1 ENST00000553802.1 ENST00000554397.1 ENST00000554919.1 ENST00000554080.1 ENST00000553371.1 |
LGMN |
legumain |
| chr21_+_35014783 | 2.59 |
ENST00000381291.4 ENST00000381285.4 ENST00000399367.3 ENST00000399352.1 ENST00000399355.2 ENST00000399349.1 |
ITSN1 |
intersectin 1 (SH3 domain protein) |
| chr8_-_49834299 | 2.56 |
ENST00000396822.1 |
SNAI2 |
snail family zinc finger 2 |
| chr16_+_69221028 | 2.50 |
ENST00000336278.4 |
SNTB2 |
syntrophin, beta 2 (dystrophin-associated protein A1, 59kDa, basic component 2) |
| chr5_+_15500280 | 2.50 |
ENST00000504595.1 |
FBXL7 |
F-box and leucine-rich repeat protein 7 |
| chr7_+_134551583 | 2.43 |
ENST00000435928.1 |
CALD1 |
caldesmon 1 |
| chr8_-_49833978 | 2.39 |
ENST00000020945.1 |
SNAI2 |
snail family zinc finger 2 |
| chr6_-_56708459 | 2.31 |
ENST00000370788.2 |
DST |
dystonin |
| chr7_-_27196267 | 2.27 |
ENST00000242159.3 |
HOXA7 |
homeobox A7 |
| chr18_-_5419797 | 2.24 |
ENST00000542146.1 ENST00000427684.2 |
EPB41L3 |
erythrocyte membrane protein band 4.1-like 3 |
| chr2_-_208030647 | 2.24 |
ENST00000309446.6 |
KLF7 |
Kruppel-like factor 7 (ubiquitous) |
| chr3_+_50284321 | 2.22 |
ENST00000451956.1 |
GNAI2 |
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2 |
| chr8_-_6420777 | 2.18 |
ENST00000415216.1 |
ANGPT2 |
angiopoietin 2 |
| chr8_-_6420930 | 2.15 |
ENST00000325203.5 |
ANGPT2 |
angiopoietin 2 |
| chr2_-_110371664 | 2.13 |
ENST00000545389.1 ENST00000423520.1 |
SEPT10 |
septin 10 |
| chr7_-_87856280 | 2.10 |
ENST00000490437.1 ENST00000431660.1 |
SRI |
sorcin |
| chr4_-_156875003 | 2.10 |
ENST00000433477.3 |
CTSO |
cathepsin O |
| chr12_+_48513570 | 2.09 |
ENST00000551804.1 |
PFKM |
phosphofructokinase, muscle |
| chr14_-_23285069 | 2.09 |
ENST00000554758.1 ENST00000397528.4 |
SLC7A7 |
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr14_-_23285011 | 2.09 |
ENST00000397532.3 |
SLC7A7 |
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr1_-_237167718 | 2.06 |
ENST00000464121.2 |
MT1HL1 |
metallothionein 1H-like 1 |
| chr1_-_23886285 | 2.03 |
ENST00000374561.5 |
ID3 |
inhibitor of DNA binding 3, dominant negative helix-loop-helix protein |
| chr15_+_32933866 | 2.01 |
ENST00000300175.4 ENST00000413748.2 ENST00000494364.1 ENST00000497208.1 |
SCG5 |
secretogranin V (7B2 protein) |
| chr5_-_125930929 | 2.00 |
ENST00000553117.1 ENST00000447989.2 ENST00000409134.3 |
ALDH7A1 |
aldehyde dehydrogenase 7 family, member A1 |
| chr17_-_46688334 | 1.98 |
ENST00000239165.7 |
HOXB7 |
homeobox B7 |
| chr17_-_33864772 | 1.93 |
ENST00000361112.4 |
SLFN12L |
schlafen family member 12-like |
| chr12_+_48513009 | 1.92 |
ENST00000359794.5 ENST00000551339.1 ENST00000395233.2 ENST00000548345.1 |
PFKM |
phosphofructokinase, muscle |
| chr14_+_77228532 | 1.84 |
ENST00000167106.4 ENST00000554237.1 |
VASH1 |
vasohibin 1 |
| chr16_+_50776021 | 1.83 |
ENST00000566679.2 ENST00000564634.1 ENST00000398568.2 |
CYLD |
cylindromatosis (turban tumor syndrome) |
| chr4_-_186697044 | 1.81 |
ENST00000437304.2 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr1_-_68698197 | 1.76 |
ENST00000370973.2 ENST00000370971.1 |
WLS |
wntless Wnt ligand secretion mediator |
| chr4_-_102268484 | 1.73 |
ENST00000394853.4 |
PPP3CA |
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr10_+_63661053 | 1.70 |
ENST00000279873.7 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
| chr10_+_31610064 | 1.70 |
ENST00000446923.2 ENST00000559476.1 |
ZEB1 |
zinc finger E-box binding homeobox 1 |
| chr1_+_39796810 | 1.69 |
ENST00000289893.4 |
MACF1 |
microtubule-actin crosslinking factor 1 |
| chr2_-_86116093 | 1.66 |
ENST00000377332.3 |
ST3GAL5 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr7_-_108096765 | 1.62 |
ENST00000379024.4 ENST00000351718.4 |
NRCAM |
neuronal cell adhesion molecule |
| chr7_-_115670804 | 1.55 |
ENST00000320239.7 |
TFEC |
transcription factor EC |
| chr5_+_66254698 | 1.54 |
ENST00000405643.1 ENST00000407621.1 ENST00000432426.1 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
| chr7_-_115670792 | 1.53 |
ENST00000265440.7 ENST00000393485.1 |
TFEC |
transcription factor EC |
| chr20_-_4795747 | 1.49 |
ENST00000379376.2 |
RASSF2 |
Ras association (RalGDS/AF-6) domain family member 2 |
| chr1_+_162602244 | 1.43 |
ENST00000367922.3 ENST00000367921.3 |
DDR2 |
discoidin domain receptor tyrosine kinase 2 |
| chr4_+_39184024 | 1.40 |
ENST00000399820.3 ENST00000509560.1 ENST00000512112.1 ENST00000288634.7 ENST00000506503.1 |
WDR19 |
WD repeat domain 19 |
| chr19_-_44124019 | 1.39 |
ENST00000300811.3 |
ZNF428 |
zinc finger protein 428 |
| chr5_+_109025067 | 1.36 |
ENST00000261483.4 |
MAN2A1 |
mannosidase, alpha, class 2A, member 1 |
| chr17_-_19281203 | 1.34 |
ENST00000487415.2 |
B9D1 |
B9 protein domain 1 |
| chr7_-_108096822 | 1.33 |
ENST00000379028.3 ENST00000413765.2 ENST00000379022.4 |
NRCAM |
neuronal cell adhesion molecule |
| chr5_+_36608422 | 1.31 |
ENST00000381918.3 |
SLC1A3 |
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr12_+_124155652 | 1.31 |
ENST00000426174.2 ENST00000303372.5 |
TCTN2 |
tectonic family member 2 |
| chr15_+_71228826 | 1.31 |
ENST00000558456.1 ENST00000560158.2 ENST00000558808.1 ENST00000559806.1 ENST00000559069.1 |
LRRC49 |
leucine rich repeat containing 49 |
| chr8_-_22926623 | 1.31 |
ENST00000276431.4 |
TNFRSF10B |
tumor necrosis factor receptor superfamily, member 10b |
| chr17_+_19281034 | 1.30 |
ENST00000308406.5 ENST00000299612.7 |
MAPK7 |
mitogen-activated protein kinase 7 |
| chr11_-_61659006 | 1.28 |
ENST00000278829.2 |
FADS3 |
fatty acid desaturase 3 |
| chr1_+_164528866 | 1.27 |
ENST00000420696.2 |
PBX1 |
pre-B-cell leukemia homeobox 1 |
| chrX_+_46696372 | 1.26 |
ENST00000218340.3 |
RP2 |
retinitis pigmentosa 2 (X-linked recessive) |
| chr8_-_6420565 | 1.25 |
ENST00000338312.6 |
ANGPT2 |
angiopoietin 2 |
| chr16_+_50775971 | 1.23 |
ENST00000311559.9 ENST00000564326.1 ENST00000566206.1 |
CYLD |
cylindromatosis (turban tumor syndrome) |
| chr6_+_153019069 | 1.20 |
ENST00000532295.1 |
MYCT1 |
myc target 1 |
| chr6_+_30457244 | 1.19 |
ENST00000376630.4 |
HLA-E |
major histocompatibility complex, class I, E |
| chr10_+_124134201 | 1.19 |
ENST00000368990.3 ENST00000368988.1 ENST00000368989.2 ENST00000463663.2 |
PLEKHA1 |
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1 |
| chr4_+_154125565 | 1.14 |
ENST00000338700.5 |
TRIM2 |
tripartite motif containing 2 |
| chr7_-_86849025 | 1.14 |
ENST00000257637.3 |
TMEM243 |
transmembrane protein 243, mitochondrial |
| chr6_-_31648150 | 1.14 |
ENST00000375858.3 ENST00000383237.4 |
LY6G5C |
lymphocyte antigen 6 complex, locus G5C |
| chr6_+_153019023 | 1.13 |
ENST00000367245.5 ENST00000529453.1 |
MYCT1 |
myc target 1 |
| chr4_-_111558135 | 1.12 |
ENST00000394598.2 ENST00000394595.3 |
PITX2 |
paired-like homeodomain 2 |
| chrX_+_49832231 | 1.11 |
ENST00000376108.3 |
CLCN5 |
chloride channel, voltage-sensitive 5 |
| chr4_-_159080806 | 1.08 |
ENST00000590648.1 |
FAM198B |
family with sequence similarity 198, member B |
| chr14_-_103523745 | 1.07 |
ENST00000361246.2 |
CDC42BPB |
CDC42 binding protein kinase beta (DMPK-like) |
| chr12_-_123752624 | 1.05 |
ENST00000542174.1 ENST00000535796.1 |
CDK2AP1 |
cyclin-dependent kinase 2 associated protein 1 |
| chr2_-_158732340 | 1.05 |
ENST00000539637.1 ENST00000413751.1 ENST00000434821.1 ENST00000424669.1 |
ACVR1 |
activin A receptor, type I |
| chr3_-_119813264 | 1.04 |
ENST00000264235.8 |
GSK3B |
glycogen synthase kinase 3 beta |
| chr15_-_34331243 | 1.03 |
ENST00000306730.3 |
AVEN |
apoptosis, caspase activation inhibitor |
| chr2_-_136873735 | 1.03 |
ENST00000409817.1 |
CXCR4 |
chemokine (C-X-C motif) receptor 4 |
| chr19_-_43708378 | 1.03 |
ENST00000599746.1 |
PSG4 |
pregnancy specific beta-1-glycoprotein 4 |
| chr17_+_8213590 | 1.02 |
ENST00000361926.3 |
ARHGEF15 |
Rho guanine nucleotide exchange factor (GEF) 15 |
| chr2_+_148602058 | 1.00 |
ENST00000241416.7 ENST00000535787.1 ENST00000404590.1 |
ACVR2A |
activin A receptor, type IIA |
| chr4_+_26585538 | 0.98 |
ENST00000264866.4 |
TBC1D19 |
TBC1 domain family, member 19 |
| chr10_+_71211212 | 0.98 |
ENST00000373290.2 |
TSPAN15 |
tetraspanin 15 |
| chr16_-_18441131 | 0.97 |
ENST00000339303.5 |
NPIPA8 |
nuclear pore complex interacting protein family, member A8 |
| chr7_+_39663061 | 0.97 |
ENST00000005257.2 |
RALA |
v-ral simian leukemia viral oncogene homolog A (ras related) |
| chr4_+_41362796 | 0.94 |
ENST00000508501.1 ENST00000512946.1 ENST00000313860.7 ENST00000512632.1 ENST00000512820.1 |
LIMCH1 |
LIM and calponin homology domains 1 |
| chr3_-_165555200 | 0.92 |
ENST00000479451.1 ENST00000540653.1 ENST00000488954.1 ENST00000264381.3 |
BCHE |
butyrylcholinesterase |
| chr3_-_158450475 | 0.92 |
ENST00000237696.5 |
RARRES1 |
retinoic acid receptor responder (tazarotene induced) 1 |
| chr19_+_41770236 | 0.91 |
ENST00000392006.3 |
HNRNPUL1 |
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr8_-_6420759 | 0.90 |
ENST00000523120.1 |
ANGPT2 |
angiopoietin 2 |
| chr15_-_59041768 | 0.86 |
ENST00000402627.1 ENST00000396140.2 ENST00000559053.1 ENST00000561288.1 |
ADAM10 |
ADAM metallopeptidase domain 10 |
| chr7_+_39663485 | 0.83 |
ENST00000436179.1 |
RALA |
v-ral simian leukemia viral oncogene homolog A (ras related) |
| chr2_-_130886795 | 0.83 |
ENST00000409914.2 |
POTEF |
POTE ankyrin domain family, member F |
| chr11_+_5617952 | 0.82 |
ENST00000354852.5 |
TRIM6-TRIM34 |
TRIM6-TRIM34 readthrough |
| chr17_+_61699766 | 0.80 |
ENST00000579585.1 ENST00000584573.1 ENST00000361733.3 ENST00000361357.3 |
MAP3K3 |
mitogen-activated protein kinase kinase kinase 3 |
| chr17_-_79481666 | 0.80 |
ENST00000575659.1 |
ACTG1 |
actin, gamma 1 |
| chr22_-_37880543 | 0.78 |
ENST00000442496.1 |
MFNG |
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr3_+_93781728 | 0.77 |
ENST00000314622.4 |
NSUN3 |
NOP2/Sun domain family, member 3 |
| chr2_+_234601512 | 0.77 |
ENST00000305139.6 |
UGT1A6 |
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr1_-_115300579 | 0.76 |
ENST00000358528.4 ENST00000525132.1 |
CSDE1 |
cold shock domain containing E1, RNA-binding |
| chrX_+_7137475 | 0.76 |
ENST00000217961.4 |
STS |
steroid sulfatase (microsomal), isozyme S |
| chr3_-_18466026 | 0.75 |
ENST00000417717.2 |
SATB1 |
SATB homeobox 1 |
| chr6_+_71122974 | 0.74 |
ENST00000418814.2 |
FAM135A |
family with sequence similarity 135, member A |
| chr1_+_220701740 | 0.73 |
ENST00000366917.4 |
MARK1 |
MAP/microtubule affinity-regulating kinase 1 |
| chr6_-_7911042 | 0.73 |
ENST00000379757.4 |
TXNDC5 |
thioredoxin domain containing 5 (endoplasmic reticulum) |
| chr18_+_11857439 | 0.72 |
ENST00000602628.1 |
GNAL |
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr16_+_50775948 | 0.71 |
ENST00000569681.1 ENST00000569418.1 ENST00000540145.1 |
CYLD |
cylindromatosis (turban tumor syndrome) |
| chr11_-_26743546 | 0.71 |
ENST00000280467.6 ENST00000396005.3 |
SLC5A12 |
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr19_+_36236514 | 0.70 |
ENST00000222266.2 |
PSENEN |
presenilin enhancer gamma secretase subunit |
| chr19_-_44123734 | 0.70 |
ENST00000598676.1 |
ZNF428 |
zinc finger protein 428 |
| chr14_+_78870030 | 0.67 |
ENST00000553631.1 ENST00000554719.1 |
NRXN3 |
neurexin 3 |
| chr2_+_201994208 | 0.67 |
ENST00000440180.1 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
| chr14_+_58765103 | 0.66 |
ENST00000355431.3 ENST00000348476.3 ENST00000395168.3 |
ARID4A |
AT rich interactive domain 4A (RBP1-like) |
| chr1_-_21377383 | 0.64 |
ENST00000374935.3 |
EIF4G3 |
eukaryotic translation initiation factor 4 gamma, 3 |
| chr13_+_111806055 | 0.63 |
ENST00000218789.5 |
ARHGEF7 |
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr15_+_67458357 | 0.63 |
ENST00000537194.2 |
SMAD3 |
SMAD family member 3 |
| chr19_-_58951496 | 0.60 |
ENST00000254166.3 |
ZNF132 |
zinc finger protein 132 |
| chr2_+_201994042 | 0.60 |
ENST00000417748.1 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
| chr19_+_41770269 | 0.59 |
ENST00000378215.4 |
HNRNPUL1 |
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr9_+_35490101 | 0.59 |
ENST00000361226.3 |
RUSC2 |
RUN and SH3 domain containing 2 |
| chr5_-_179334825 | 0.59 |
ENST00000355235.3 ENST00000356834.3 |
TBC1D9B |
TBC1 domain family, member 9B (with GRAM domain) |
| chr17_+_61554413 | 0.58 |
ENST00000538928.1 ENST00000290866.4 ENST00000428043.1 |
ACE |
angiotensin I converting enzyme |
| chr22_+_38864041 | 0.58 |
ENST00000216014.4 ENST00000409006.3 |
KDELR3 |
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
| chr7_+_149535455 | 0.58 |
ENST00000223210.4 ENST00000460379.1 |
ZNF862 |
zinc finger protein 862 |
| chr11_+_93861993 | 0.58 |
ENST00000227638.3 ENST00000436171.2 |
PANX1 |
pannexin 1 |
| chr16_+_68279207 | 0.56 |
ENST00000413021.2 ENST00000565744.1 ENST00000219345.5 |
PLA2G15 |
phospholipase A2, group XV |
| chr5_-_137071756 | 0.56 |
ENST00000394937.3 ENST00000309755.4 |
KLHL3 |
kelch-like family member 3 |
| chr5_-_137475071 | 0.56 |
ENST00000265191.2 |
NME5 |
NME/NM23 family member 5 |
| chr3_-_158450231 | 0.56 |
ENST00000479756.1 |
RARRES1 |
retinoic acid receptor responder (tazarotene induced) 1 |
| chr10_+_72575643 | 0.55 |
ENST00000373202.3 |
SGPL1 |
sphingosine-1-phosphate lyase 1 |
| chr4_-_186696425 | 0.55 |
ENST00000430503.1 ENST00000319454.6 ENST00000450341.1 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr6_-_116601044 | 0.55 |
ENST00000368608.3 |
TSPYL1 |
TSPY-like 1 |
| chr2_-_89160770 | 0.55 |
ENST00000390240.2 |
IGKJ3 |
immunoglobulin kappa joining 3 |
| chr6_+_25963020 | 0.55 |
ENST00000357085.3 |
TRIM38 |
tripartite motif containing 38 |
| chr1_-_115300592 | 0.53 |
ENST00000261443.5 ENST00000534699.1 ENST00000339438.6 ENST00000529046.1 ENST00000525970.1 ENST00000369530.1 ENST00000530886.1 |
CSDE1 |
cold shock domain containing E1, RNA-binding |
| chr9_-_95640218 | 0.52 |
ENST00000395506.3 ENST00000375495.3 ENST00000332591.6 |
ZNF484 |
zinc finger protein 484 |
| chr16_-_18468926 | 0.51 |
ENST00000545114.1 |
RP11-1212A22.4 |
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chr6_+_90272027 | 0.50 |
ENST00000522441.1 |
ANKRD6 |
ankyrin repeat domain 6 |
| chr2_-_9563469 | 0.49 |
ENST00000484735.1 ENST00000456913.2 |
ITGB1BP1 |
integrin beta 1 binding protein 1 |
| chr1_-_230513367 | 0.49 |
ENST00000321327.2 ENST00000525115.1 |
PGBD5 |
piggyBac transposable element derived 5 |
| chr1_-_21377447 | 0.48 |
ENST00000374937.3 ENST00000264211.8 |
EIF4G3 |
eukaryotic translation initiation factor 4 gamma, 3 |
| chr19_+_36236491 | 0.48 |
ENST00000591949.1 |
PSENEN |
presenilin enhancer gamma secretase subunit |
| chr18_-_48723690 | 0.48 |
ENST00000406189.3 |
MEX3C |
mex-3 RNA binding family member C |
| chr11_+_74699703 | 0.48 |
ENST00000529024.1 ENST00000544263.1 |
NEU3 |
sialidase 3 (membrane sialidase) |
| chr21_+_47743995 | 0.47 |
ENST00000359568.5 |
PCNT |
pericentrin |
| chr14_-_80677815 | 0.46 |
ENST00000557125.1 ENST00000555750.1 |
DIO2 |
deiodinase, iodothyronine, type II |
| chr2_-_45236540 | 0.44 |
ENST00000303077.6 |
SIX2 |
SIX homeobox 2 |
| chr9_+_35538616 | 0.43 |
ENST00000455600.1 |
RUSC2 |
RUN and SH3 domain containing 2 |
| chr14_-_80677970 | 0.42 |
ENST00000438257.4 |
DIO2 |
deiodinase, iodothyronine, type II |
| chr19_-_4831701 | 0.42 |
ENST00000248244.5 |
TICAM1 |
toll-like receptor adaptor molecule 1 |
| chr3_+_173116225 | 0.42 |
ENST00000457714.1 |
NLGN1 |
neuroligin 1 |
| chr2_+_242295658 | 0.40 |
ENST00000264042.3 ENST00000545004.1 ENST00000373287.4 |
FARP2 |
FERM, RhoGEF and pleckstrin domain protein 2 |
| chr3_-_195619579 | 0.40 |
ENST00000428187.1 |
TNK2 |
tyrosine kinase, non-receptor, 2 |
| chr2_-_9563319 | 0.39 |
ENST00000497105.1 ENST00000360635.3 ENST00000359712.3 |
ITGB1BP1 |
integrin beta 1 binding protein 1 |
| chr10_-_49813090 | 0.39 |
ENST00000249601.4 |
ARHGAP22 |
Rho GTPase activating protein 22 |
| chr19_+_3933085 | 0.38 |
ENST00000168977.2 ENST00000599576.1 |
NMRK2 |
nicotinamide riboside kinase 2 |
| chr6_+_26440700 | 0.37 |
ENST00000494393.1 ENST00000482451.1 ENST00000244519.2 ENST00000339789.4 ENST00000471353.1 ENST00000361232.3 ENST00000487627.1 ENST00000496719.1 ENST00000490254.1 ENST00000487272.1 |
BTN3A3 |
butyrophilin, subfamily 3, member A3 |
| chr19_-_4457776 | 0.37 |
ENST00000301281.6 |
UBXN6 |
UBX domain protein 6 |
| chr3_-_15540055 | 0.36 |
ENST00000605797.1 ENST00000435459.2 |
COLQ |
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
| chr19_-_43269809 | 0.36 |
ENST00000406636.3 ENST00000404209.4 ENST00000306511.4 |
PSG8 |
pregnancy specific beta-1-glycoprotein 8 |
| chr11_+_30253410 | 0.35 |
ENST00000533718.1 |
FSHB |
follicle stimulating hormone, beta polypeptide |
| chr19_+_41770349 | 0.35 |
ENST00000602130.1 |
HNRNPUL1 |
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr8_-_59412717 | 0.35 |
ENST00000301645.3 |
CYP7A1 |
cytochrome P450, family 7, subfamily A, polypeptide 1 |
| chr6_+_71123107 | 0.34 |
ENST00000370479.3 ENST00000505769.1 ENST00000515323.1 ENST00000515280.1 ENST00000507085.1 ENST00000457062.2 ENST00000361499.3 |
FAM135A |
family with sequence similarity 135, member A |
| chr11_-_82746587 | 0.34 |
ENST00000528379.1 ENST00000534103.1 |
RAB30 |
RAB30, member RAS oncogene family |
| chr19_-_10687907 | 0.34 |
ENST00000589348.1 |
AP1M2 |
adaptor-related protein complex 1, mu 2 subunit |
| chr20_+_36974759 | 0.34 |
ENST00000217407.2 |
LBP |
lipopolysaccharide binding protein |
| chr11_-_33774944 | 0.33 |
ENST00000532057.1 ENST00000531080.1 |
FBXO3 |
F-box protein 3 |
| chr5_+_140529630 | 0.33 |
ENST00000543635.1 |
PCDHB6 |
protocadherin beta 6 |
| chr6_-_131321863 | 0.32 |
ENST00000528282.1 |
EPB41L2 |
erythrocyte membrane protein band 4.1-like 2 |
| chr20_-_57607347 | 0.32 |
ENST00000395663.1 ENST00000395659.1 ENST00000243997.3 |
ATP5E |
ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit |
| chrX_+_84499081 | 0.32 |
ENST00000276123.3 |
ZNF711 |
zinc finger protein 711 |
| chr5_+_14143728 | 0.31 |
ENST00000344204.4 ENST00000537187.1 |
TRIO |
trio Rho guanine nucleotide exchange factor |
| chr17_-_79827808 | 0.30 |
ENST00000580685.1 |
ARHGDIA |
Rho GDP dissociation inhibitor (GDI) alpha |
| chr10_-_129924468 | 0.30 |
ENST00000368653.3 |
MKI67 |
marker of proliferation Ki-67 |
| chr2_-_85788652 | 0.30 |
ENST00000430215.3 |
GGCX |
gamma-glutamyl carboxylase |
| chr19_+_41103063 | 0.30 |
ENST00000308370.7 |
LTBP4 |
latent transforming growth factor beta binding protein 4 |
| chr10_+_97471508 | 0.30 |
ENST00000453258.2 |
ENTPD1 |
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr6_-_35765088 | 0.30 |
ENST00000259938.2 |
CLPS |
colipase, pancreatic |
| chr15_-_42500351 | 0.29 |
ENST00000348544.4 ENST00000318006.5 |
VPS39 |
vacuolar protein sorting 39 homolog (S. cerevisiae) |
| chr4_+_56815102 | 0.28 |
ENST00000257287.4 |
CEP135 |
centrosomal protein 135kDa |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.7 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 1.8 | 5.5 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 1.7 | 5.1 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 1.6 | 4.9 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 1.6 | 6.5 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 1.3 | 4.0 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 1.3 | 3.8 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 1.1 | 12.8 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 1.1 | 5.3 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 1.0 | 2.9 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) |
| 0.9 | 4.5 | GO:0003289 | atrial septum primum morphogenesis(GO:0003289) |
| 0.9 | 6.1 | GO:2000795 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.8 | 3.1 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 0.7 | 4.4 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.6 | 1.8 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.6 | 5.3 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.6 | 1.7 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.5 | 2.9 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.5 | 1.4 | GO:0061055 | myotome development(GO:0061055) |
| 0.5 | 2.3 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.5 | 5.1 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.4 | 2.7 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.4 | 1.2 | GO:0032679 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) TRAIL production(GO:0032639) regulation of TRAIL production(GO:0032679) positive regulation of TRAIL production(GO:0032759) protection from natural killer cell mediated cytotoxicity(GO:0042270) positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.4 | 2.2 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.4 | 1.1 | GO:0021763 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) left lung development(GO:0060459) left lung morphogenesis(GO:0060460) pulmonary vein morphogenesis(GO:0060577) superior vena cava morphogenesis(GO:0060578) |
| 0.3 | 3.7 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.3 | 0.9 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.3 | 2.1 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.3 | 0.9 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.3 | 2.0 | GO:0042426 | choline catabolic process(GO:0042426) |
| 0.3 | 0.8 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.3 | 2.3 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.2 | 2.5 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.2 | 6.2 | GO:0033622 | integrin activation(GO:0033622) |
| 0.2 | 1.3 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.2 | 1.3 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.2 | 1.7 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.2 | 1.7 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.2 | 1.3 | GO:0006537 | glutamate biosynthetic process(GO:0006537) gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.2 | 2.5 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.2 | 1.4 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.2 | 1.2 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.2 | 0.7 | GO:0097368 | histone H4-K20 trimethylation(GO:0034773) establishment of Sertoli cell barrier(GO:0097368) |
| 0.2 | 1.7 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.2 | 1.8 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.1 | 0.4 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.1 | 1.0 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.1 | 0.6 | GO:1903597 | negative regulation of gap junction assembly(GO:1903597) |
| 0.1 | 0.6 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.1 | 0.4 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.1 | 3.3 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 | 1.3 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.1 | 0.6 | GO:1903243 | negative regulation of cardiac muscle adaptation(GO:0010616) negative regulation of lung blood pressure(GO:0061767) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.1 | 0.3 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.1 | 0.3 | GO:0015920 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) lipopolysaccharide transport(GO:0015920) |
| 0.1 | 1.4 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 | 1.2 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.1 | 1.0 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 | 0.3 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 0.4 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.1 | 0.6 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.9 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 0.5 | GO:0003415 | chondrocyte hypertrophy(GO:0003415) |
| 0.1 | 0.7 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.1 | 1.2 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.1 | 2.4 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 0.3 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.1 | 0.8 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 0.6 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.1 | 0.2 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 0.2 | GO:1900114 | positive regulation of histone H3-K9 trimethylation(GO:1900114) |
| 0.1 | 0.4 | GO:0045359 | negative regulation of MyD88-independent toll-like receptor signaling pathway(GO:0034128) positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.1 | 0.8 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 0.5 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 | 1.3 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 0.2 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.1 | 0.6 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 0.3 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.1 | 0.2 | GO:0006524 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) |
| 0.1 | 0.2 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.1 | 0.5 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 2.0 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.1 | GO:0032887 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 | 2.0 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 1.7 | GO:0016577 | histone demethylation(GO:0016577) |
| 0.0 | 0.8 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 1.1 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.0 | 0.4 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.0 | 2.0 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.6 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.8 | GO:0006706 | steroid catabolic process(GO:0006706) |
| 0.0 | 0.7 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.5 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.3 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.8 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 0.7 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 1.3 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.7 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.6 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.1 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.0 | 0.2 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.1 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) formation of anatomical boundary(GO:0048859) cell proliferation involved in heart valve development(GO:2000793) |
| 0.0 | 0.2 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.0 | 1.1 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.0 | 0.2 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.0 | 0.4 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 1.6 | GO:0001895 | retina homeostasis(GO:0001895) |
| 0.0 | 0.4 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 2.7 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 0.3 | GO:0044241 | lipid digestion(GO:0044241) |
| 0.0 | 1.4 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.4 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 1.4 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.3 | GO:0034656 | nucleobase-containing small molecule catabolic process(GO:0034656) |
| 0.0 | 0.2 | GO:0060117 | auditory receptor cell development(GO:0060117) |
| 0.0 | 0.9 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.0 | 1.0 | GO:0008593 | regulation of Notch signaling pathway(GO:0008593) |
| 0.0 | 0.3 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.3 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.4 | GO:0050687 | negative regulation of defense response to virus(GO:0050687) |
| 0.0 | 1.1 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 6.9 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 1.8 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.3 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 0.3 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.0 | 0.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.5 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.4 | 6.6 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.2 | 12.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.2 | 1.0 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.2 | 6.5 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.2 | 6.1 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.2 | 4.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 10.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 2.0 | REACTOME SIGNALING BY NOTCH2 | Genes involved in Signaling by NOTCH2 |
| 0.1 | 4.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 1.8 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 4.2 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 2.6 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 4.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 3.8 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.1 | 2.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 0.4 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 0.6 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 0.8 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 1.7 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 3.3 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.1 | 1.3 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 0.9 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 3.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.8 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 1.1 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 1.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.7 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 5.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.8 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.5 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 1.1 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 4.7 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.4 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.8 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 2.2 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.3 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 2.0 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.2 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.0 | 0.8 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.3 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 1.1 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.5 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 1.0 | 5.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.9 | 5.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.7 | 4.0 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.4 | 5.7 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.4 | 4.2 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.4 | 3.8 | GO:1990380 | Lys63-specific deubiquitinase activity(GO:0061578) Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.4 | 3.7 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.3 | 8.9 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.3 | 2.1 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.3 | 1.4 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.3 | 12.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 2.9 | GO:0046625 | sphingosine-1-phosphate receptor activity(GO:0038036) sphingolipid binding(GO:0046625) |
| 0.2 | 0.9 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.2 | 2.9 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.2 | 1.3 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.2 | 0.8 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.2 | 0.6 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.2 | 1.3 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.2 | 2.7 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.2 | 1.7 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.2 | 2.9 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.2 | 4.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 0.8 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.1 | 1.3 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.1 | 1.2 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.9 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 5.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 1.2 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.1 | 1.7 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.6 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 0.5 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.1 | 0.2 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.1 | 1.4 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.1 | 4.9 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 0.6 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) peptidyl-dipeptidase activity(GO:0008241) |
| 0.1 | 2.0 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 0.4 | GO:0061769 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.1 | 1.0 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 0.3 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.1 | 0.3 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 2.0 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 2.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 6.5 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 2.0 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 4.5 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 0.7 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 10.0 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 0.6 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 1.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.6 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.1 | 1.3 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 0.2 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 0.2 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.1 | 0.5 | GO:0008233 | peptidase activity(GO:0008233) |
| 0.1 | 0.2 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.1 | 1.7 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.1 | 1.1 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.1 | 0.2 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.1 | 4.9 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 1.1 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.9 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 1.2 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.3 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.7 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.3 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.0 | 0.5 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 1.1 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.8 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 14.0 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.3 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 1.7 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 2.5 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.6 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.5 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 1.9 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 2.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.6 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 8.6 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.3 | GO:0016831 | carbon-carbon lyase activity(GO:0016830) carboxy-lyase activity(GO:0016831) |
| 0.0 | 2.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.3 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 1.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.1 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.3 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.0 | 0.5 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.2 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.7 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.9 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.1 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.3 | GO:0031673 | H zone(GO:0031673) |
| 1.0 | 3.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.8 | 4.0 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.8 | 5.5 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.7 | 3.0 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.7 | 10.4 | GO:0030478 | actin cap(GO:0030478) |
| 0.7 | 2.7 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.5 | 6.1 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.3 | 1.4 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.3 | 1.3 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.3 | 2.1 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.3 | 1.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.2 | 1.4 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.2 | 5.7 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.2 | 1.7 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.2 | 2.7 | GO:0036038 | MKS complex(GO:0036038) |
| 0.2 | 1.8 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.2 | 2.9 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 2.2 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 4.9 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 12.8 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 1.3 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 6.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 1.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 1.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 3.8 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 1.0 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 0.9 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 0.6 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 0.6 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 2.5 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 0.3 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 0.4 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.1 | 0.8 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 1.1 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.1 | 1.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0061673 | cortical microtubule(GO:0055028) mitotic spindle astral microtubule(GO:0061673) |
| 0.0 | 0.3 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 2.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.3 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 4.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.2 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) GID complex(GO:0034657) |
| 0.0 | 0.6 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.4 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.3 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 5.1 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 1.8 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.8 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.6 | GO:0036126 | sperm flagellum(GO:0036126) sperm part(GO:0097223) |
| 0.0 | 2.9 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.2 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.1 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.0 | 2.7 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 0.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 1.3 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.9 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 3.3 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 3.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 9.1 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.2 | 13.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 3.8 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 2.6 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.1 | 1.0 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.1 | 6.5 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 6.0 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 5.8 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 2.6 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 3.1 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 2.0 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 1.1 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 1.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 4.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 4.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 5.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 1.0 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.1 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 1.9 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 1.2 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 1.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 5.5 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.0 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 2.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 3.1 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 7.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.6 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 1.1 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.1 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.3 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.2 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.3 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.4 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |