Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
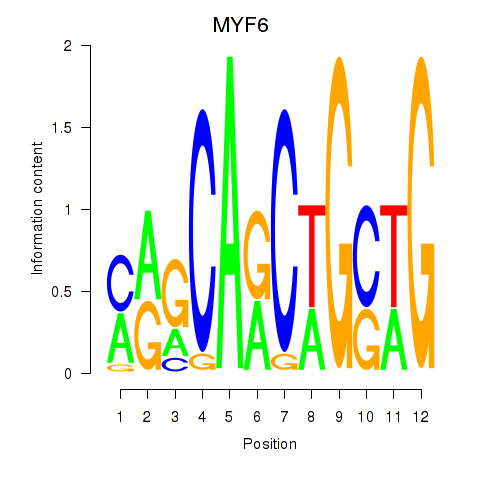
Results for MYF6
Z-value: 1.39
Transcription factors associated with MYF6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MYF6
|
ENSG00000111046.3 | MYF6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MYF6 | hg19_v2_chr12_+_81101277_81101321 | -0.10 | 7.3e-01 | Click! |
Activity profile of MYF6 motif
Sorted Z-values of MYF6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MYF6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_53253112 | 4.82 |
ENST00000568673.1 ENST00000562847.1 ENST00000568147.1 |
TCF4 |
transcription factor 4 |
| chrX_-_38080077 | 4.80 |
ENST00000378533.3 ENST00000544439.1 ENST00000432886.2 ENST00000538295.1 |
SRPX |
sushi-repeat containing protein, X-linked |
| chr18_-_53253323 | 4.49 |
ENST00000540999.1 ENST00000563888.2 |
TCF4 |
transcription factor 4 |
| chr9_+_116298778 | 3.59 |
ENST00000462143.1 |
RGS3 |
regulator of G-protein signaling 3 |
| chr11_+_114168773 | 2.96 |
ENST00000542647.1 ENST00000545255.1 |
NNMT |
nicotinamide N-methyltransferase |
| chr13_-_72440901 | 2.43 |
ENST00000359684.2 |
DACH1 |
dachshund homolog 1 (Drosophila) |
| chr13_-_108518986 | 2.33 |
ENST00000375915.2 |
FAM155A |
family with sequence similarity 155, member A |
| chr1_-_57045228 | 2.08 |
ENST00000371250.3 |
PPAP2B |
phosphatidic acid phosphatase type 2B |
| chr5_-_94620239 | 1.97 |
ENST00000515393.1 |
MCTP1 |
multiple C2 domains, transmembrane 1 |
| chr10_+_89419370 | 1.91 |
ENST00000361175.4 ENST00000456849.1 |
PAPSS2 |
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr3_-_124774802 | 1.90 |
ENST00000311127.4 |
HEG1 |
heart development protein with EGF-like domains 1 |
| chr1_-_156647189 | 1.90 |
ENST00000368223.3 |
NES |
nestin |
| chr15_+_96873921 | 1.76 |
ENST00000394166.3 |
NR2F2 |
nuclear receptor subfamily 2, group F, member 2 |
| chr6_+_7727030 | 1.63 |
ENST00000283147.6 |
BMP6 |
bone morphogenetic protein 6 |
| chr3_-_145878954 | 1.60 |
ENST00000282903.5 ENST00000360060.3 |
PLOD2 |
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 |
| chr20_-_23066953 | 1.56 |
ENST00000246006.4 |
CD93 |
CD93 molecule |
| chr2_-_161349909 | 1.51 |
ENST00000392753.3 |
RBMS1 |
RNA binding motif, single stranded interacting protein 1 |
| chr9_-_35685452 | 1.49 |
ENST00000607559.1 |
TPM2 |
tropomyosin 2 (beta) |
| chr17_+_68165657 | 1.47 |
ENST00000243457.3 |
KCNJ2 |
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr1_+_150245099 | 1.41 |
ENST00000369099.3 |
C1orf54 |
chromosome 1 open reading frame 54 |
| chr9_+_137533615 | 1.40 |
ENST00000371817.3 |
COL5A1 |
collagen, type V, alpha 1 |
| chr7_-_27183263 | 1.40 |
ENST00000222726.3 |
HOXA5 |
homeobox A5 |
| chr11_-_86666427 | 1.35 |
ENST00000531380.1 |
FZD4 |
frizzled family receptor 4 |
| chr21_+_37507210 | 1.34 |
ENST00000290354.5 |
CBR3 |
carbonyl reductase 3 |
| chr2_+_37571717 | 1.33 |
ENST00000338415.3 ENST00000404976.1 |
QPCT |
glutaminyl-peptide cyclotransferase |
| chr2_-_110371720 | 1.32 |
ENST00000356688.4 |
SEPT10 |
septin 10 |
| chr2_+_37571845 | 1.32 |
ENST00000537448.1 |
QPCT |
glutaminyl-peptide cyclotransferase |
| chr7_+_102553430 | 1.24 |
ENST00000339431.4 ENST00000249377.4 |
LRRC17 |
leucine rich repeat containing 17 |
| chr20_+_33759854 | 1.22 |
ENST00000216968.4 |
PROCR |
protein C receptor, endothelial |
| chr12_-_12503156 | 1.22 |
ENST00000543314.1 ENST00000396349.3 |
MANSC1 |
MANSC domain containing 1 |
| chrX_+_100333709 | 1.22 |
ENST00000372930.4 |
TMEM35 |
transmembrane protein 35 |
| chr12_-_54813229 | 1.22 |
ENST00000293379.4 |
ITGA5 |
integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
| chr5_-_16936340 | 1.20 |
ENST00000507288.1 ENST00000513610.1 |
MYO10 |
myosin X |
| chr4_-_52904425 | 1.17 |
ENST00000535450.1 |
SGCB |
sarcoglycan, beta (43kDa dystrophin-associated glycoprotein) |
| chr8_-_27472198 | 1.16 |
ENST00000519472.1 ENST00000523589.1 ENST00000522413.1 ENST00000523396.1 ENST00000560366.1 |
CLU |
clusterin |
| chr3_+_110790590 | 1.13 |
ENST00000485303.1 |
PVRL3 |
poliovirus receptor-related 3 |
| chr14_-_54423529 | 1.12 |
ENST00000245451.4 ENST00000559087.1 |
BMP4 |
bone morphogenetic protein 4 |
| chr19_+_48216600 | 1.11 |
ENST00000263277.3 ENST00000538399.1 |
EHD2 |
EH-domain containing 2 |
| chr7_+_100199800 | 1.10 |
ENST00000223061.5 |
PCOLCE |
procollagen C-endopeptidase enhancer |
| chr7_-_11871815 | 1.10 |
ENST00000423059.4 |
THSD7A |
thrombospondin, type I, domain containing 7A |
| chr2_+_217498105 | 1.10 |
ENST00000233809.4 |
IGFBP2 |
insulin-like growth factor binding protein 2, 36kDa |
| chr5_+_140864649 | 1.09 |
ENST00000306593.1 |
PCDHGC4 |
protocadherin gamma subfamily C, 4 |
| chr12_-_6665200 | 1.08 |
ENST00000336604.4 ENST00000396840.2 ENST00000356896.4 |
IFFO1 |
intermediate filament family orphan 1 |
| chr17_+_39240459 | 1.06 |
ENST00000391417.4 |
KRTAP4-7 |
keratin associated protein 4-7 |
| chr3_-_129325660 | 1.06 |
ENST00000324093.4 ENST00000393239.1 |
PLXND1 |
plexin D1 |
| chr12_-_49318715 | 1.00 |
ENST00000444214.2 |
FKBP11 |
FK506 binding protein 11, 19 kDa |
| chr10_+_129845823 | 1.00 |
ENST00000306042.5 |
PTPRE |
protein tyrosine phosphatase, receptor type, E |
| chr12_-_49319265 | 0.99 |
ENST00000552878.1 ENST00000453172.2 |
FKBP11 |
FK506 binding protein 11, 19 kDa |
| chr1_+_25071848 | 0.98 |
ENST00000374379.4 |
CLIC4 |
chloride intracellular channel 4 |
| chr4_+_74735102 | 0.97 |
ENST00000395761.3 |
CXCL1 |
chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) |
| chr1_+_180123969 | 0.95 |
ENST00000367602.3 ENST00000367600.5 |
QSOX1 |
quiescin Q6 sulfhydryl oxidase 1 |
| chr2_-_216300784 | 0.95 |
ENST00000421182.1 ENST00000432072.2 ENST00000323926.6 ENST00000336916.4 ENST00000357867.4 ENST00000359671.1 ENST00000346544.3 ENST00000345488.5 ENST00000357009.2 ENST00000446046.1 ENST00000356005.4 ENST00000443816.1 ENST00000426059.1 ENST00000354785.4 |
FN1 |
fibronectin 1 |
| chr22_+_38071615 | 0.94 |
ENST00000215909.5 |
LGALS1 |
lectin, galactoside-binding, soluble, 1 |
| chr6_-_7911042 | 0.94 |
ENST00000379757.4 |
TXNDC5 |
thioredoxin domain containing 5 (endoplasmic reticulum) |
| chr8_-_23712312 | 0.94 |
ENST00000290271.2 |
STC1 |
stanniocalcin 1 |
| chrX_-_140271249 | 0.92 |
ENST00000370526.2 |
LDOC1 |
leucine zipper, down-regulated in cancer 1 |
| chr1_-_225840747 | 0.91 |
ENST00000366843.2 ENST00000366844.3 |
ENAH |
enabled homolog (Drosophila) |
| chr4_-_5021164 | 0.89 |
ENST00000506508.1 ENST00000509419.1 ENST00000307746.4 |
CYTL1 |
cytokine-like 1 |
| chr6_+_123110465 | 0.88 |
ENST00000539041.1 |
SMPDL3A |
sphingomyelin phosphodiesterase, acid-like 3A |
| chr6_+_123110302 | 0.87 |
ENST00000368440.4 |
SMPDL3A |
sphingomyelin phosphodiesterase, acid-like 3A |
| chr7_-_137028534 | 0.87 |
ENST00000348225.2 |
PTN |
pleiotrophin |
| chr6_-_167275991 | 0.86 |
ENST00000510118.1 |
RPS6KA2 |
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
| chr2_-_110371777 | 0.86 |
ENST00000397712.2 |
SEPT10 |
septin 10 |
| chr4_+_47487285 | 0.86 |
ENST00000273859.3 ENST00000504445.1 |
ATP10D |
ATPase, class V, type 10D |
| chr1_-_236228417 | 0.86 |
ENST00000264187.6 |
NID1 |
nidogen 1 |
| chr7_-_137028498 | 0.84 |
ENST00000393083.2 |
PTN |
pleiotrophin |
| chr7_-_94285472 | 0.84 |
ENST00000437425.2 ENST00000447873.1 ENST00000415788.2 |
SGCE |
sarcoglycan, epsilon |
| chr10_-_90967063 | 0.83 |
ENST00000371852.2 |
CH25H |
cholesterol 25-hydroxylase |
| chrX_+_54835493 | 0.83 |
ENST00000396224.1 |
MAGED2 |
melanoma antigen family D, 2 |
| chr3_-_49395892 | 0.82 |
ENST00000419783.1 |
GPX1 |
glutathione peroxidase 1 |
| chr7_+_94023873 | 0.82 |
ENST00000297268.6 |
COL1A2 |
collagen, type I, alpha 2 |
| chr13_-_24007815 | 0.82 |
ENST00000382298.3 |
SACS |
spastic ataxia of Charlevoix-Saguenay (sacsin) |
| chr7_-_94285511 | 0.82 |
ENST00000265735.7 |
SGCE |
sarcoglycan, epsilon |
| chr1_-_236228403 | 0.81 |
ENST00000366595.3 |
NID1 |
nidogen 1 |
| chr6_-_32191834 | 0.81 |
ENST00000375023.3 |
NOTCH4 |
notch 4 |
| chr12_-_71003568 | 0.81 |
ENST00000547715.1 ENST00000451516.2 ENST00000538708.1 ENST00000550857.1 ENST00000261266.5 |
PTPRB |
protein tyrosine phosphatase, receptor type, B |
| chr20_-_57582296 | 0.78 |
ENST00000217131.5 |
CTSZ |
cathepsin Z |
| chr16_-_74640986 | 0.78 |
ENST00000422840.2 ENST00000565260.1 ENST00000447066.2 ENST00000205061.5 |
GLG1 |
golgi glycoprotein 1 |
| chr3_-_46037299 | 0.77 |
ENST00000296137.2 |
FYCO1 |
FYVE and coiled-coil domain containing 1 |
| chr17_-_39968855 | 0.77 |
ENST00000355468.3 ENST00000590496.1 |
LEPREL4 |
leprecan-like 4 |
| chr15_+_96875657 | 0.76 |
ENST00000559679.1 ENST00000394171.2 |
NR2F2 |
nuclear receptor subfamily 2, group F, member 2 |
| chr14_+_24867992 | 0.76 |
ENST00000382554.3 |
NYNRIN |
NYN domain and retroviral integrase containing |
| chr11_-_66336060 | 0.76 |
ENST00000310325.5 |
CTSF |
cathepsin F |
| chr2_-_110371412 | 0.75 |
ENST00000415095.1 ENST00000334001.6 ENST00000437928.1 ENST00000493445.1 ENST00000397714.2 ENST00000461295.1 |
SEPT10 |
septin 10 |
| chr11_-_119187826 | 0.75 |
ENST00000264036.4 |
MCAM |
melanoma cell adhesion molecule |
| chr1_+_153747746 | 0.74 |
ENST00000368661.3 |
SLC27A3 |
solute carrier family 27 (fatty acid transporter), member 3 |
| chr19_+_35521572 | 0.73 |
ENST00000262631.5 |
SCN1B |
sodium channel, voltage-gated, type I, beta subunit |
| chr1_+_180165672 | 0.73 |
ENST00000443059.1 |
QSOX1 |
quiescin Q6 sulfhydryl oxidase 1 |
| chr7_-_151433342 | 0.73 |
ENST00000433631.2 |
PRKAG2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr22_+_31489344 | 0.73 |
ENST00000404574.1 |
SMTN |
smoothelin |
| chr9_-_13165457 | 0.72 |
ENST00000542239.1 ENST00000538841.1 ENST00000433359.2 |
MPDZ |
multiple PDZ domain protein |
| chr11_+_128634589 | 0.72 |
ENST00000281428.8 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
| chr7_-_124405681 | 0.72 |
ENST00000303921.2 |
GPR37 |
G protein-coupled receptor 37 (endothelin receptor type B-like) |
| chr8_-_62627057 | 0.71 |
ENST00000519234.1 ENST00000379449.6 ENST00000379454.4 ENST00000518068.1 ENST00000517856.1 ENST00000356457.5 |
ASPH |
aspartate beta-hydroxylase |
| chr8_+_40010989 | 0.70 |
ENST00000315792.3 |
C8orf4 |
chromosome 8 open reading frame 4 |
| chrX_-_119694538 | 0.70 |
ENST00000371322.5 |
CUL4B |
cullin 4B |
| chr17_-_1549011 | 0.69 |
ENST00000571272.1 ENST00000263071.4 ENST00000348987.3 |
SCARF1 |
scavenger receptor class F, member 1 |
| chr11_-_117166276 | 0.69 |
ENST00000510630.1 ENST00000392937.6 |
BACE1 |
beta-site APP-cleaving enzyme 1 |
| chr14_-_54421190 | 0.69 |
ENST00000417573.1 |
BMP4 |
bone morphogenetic protein 4 |
| chr21_+_46875395 | 0.69 |
ENST00000355480.5 |
COL18A1 |
collagen, type XVIII, alpha 1 |
| chr1_+_51701924 | 0.68 |
ENST00000242719.3 |
RNF11 |
ring finger protein 11 |
| chr22_-_37882395 | 0.68 |
ENST00000416983.3 ENST00000424765.2 ENST00000356998.3 |
MFNG |
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr14_+_58765103 | 0.68 |
ENST00000355431.3 ENST00000348476.3 ENST00000395168.3 |
ARID4A |
AT rich interactive domain 4A (RBP1-like) |
| chr1_-_227505289 | 0.68 |
ENST00000366765.3 |
CDC42BPA |
CDC42 binding protein kinase alpha (DMPK-like) |
| chr13_-_114567034 | 0.67 |
ENST00000327773.6 ENST00000357389.3 |
GAS6 |
growth arrest-specific 6 |
| chr19_-_19051103 | 0.67 |
ENST00000542541.2 ENST00000433218.2 |
HOMER3 |
homer homolog 3 (Drosophila) |
| chr14_-_75422280 | 0.67 |
ENST00000238607.6 ENST00000553716.1 |
PGF |
placental growth factor |
| chr9_+_116225999 | 0.66 |
ENST00000317613.6 |
RGS3 |
regulator of G-protein signaling 3 |
| chr21_+_46875424 | 0.66 |
ENST00000359759.4 |
COL18A1 |
collagen, type XVIII, alpha 1 |
| chr7_-_27169801 | 0.65 |
ENST00000511914.1 |
HOXA4 |
homeobox A4 |
| chr4_-_102268484 | 0.65 |
ENST00000394853.4 |
PPP3CA |
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr1_+_23037323 | 0.65 |
ENST00000544305.1 ENST00000374630.3 ENST00000400191.3 ENST00000374632.3 |
EPHB2 |
EPH receptor B2 |
| chr9_+_125133315 | 0.64 |
ENST00000223423.4 ENST00000362012.2 |
PTGS1 |
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr4_+_15704679 | 0.64 |
ENST00000382346.3 |
BST1 |
bone marrow stromal cell antigen 1 |
| chr3_-_99833333 | 0.63 |
ENST00000354552.3 ENST00000331335.5 ENST00000398326.2 |
FILIP1L |
filamin A interacting protein 1-like |
| chr3_-_69062764 | 0.63 |
ENST00000295571.5 |
EOGT |
EGF domain-specific O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr6_-_31324943 | 0.62 |
ENST00000412585.2 ENST00000434333.1 |
HLA-B |
major histocompatibility complex, class I, B |
| chr19_-_44174305 | 0.61 |
ENST00000601723.1 ENST00000339082.3 |
PLAUR |
plasminogen activator, urokinase receptor |
| chr12_-_15103621 | 0.61 |
ENST00000536592.1 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
| chr3_-_87040233 | 0.60 |
ENST00000398399.2 |
VGLL3 |
vestigial like 3 (Drosophila) |
| chr1_+_66458072 | 0.60 |
ENST00000423207.2 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
| chrX_-_153637612 | 0.59 |
ENST00000369807.1 ENST00000369808.3 |
DNASE1L1 |
deoxyribonuclease I-like 1 |
| chr7_-_151433393 | 0.58 |
ENST00000492843.1 |
PRKAG2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chrX_+_23685563 | 0.57 |
ENST00000379341.4 |
PRDX4 |
peroxiredoxin 4 |
| chr3_+_45067659 | 0.57 |
ENST00000296130.4 |
CLEC3B |
C-type lectin domain family 3, member B |
| chr12_-_56122761 | 0.57 |
ENST00000552164.1 ENST00000420846.3 ENST00000257857.4 |
CD63 |
CD63 molecule |
| chr14_+_90863327 | 0.55 |
ENST00000356978.4 |
CALM1 |
calmodulin 1 (phosphorylase kinase, delta) |
| chr10_-_62704005 | 0.55 |
ENST00000337910.5 |
RHOBTB1 |
Rho-related BTB domain containing 1 |
| chr19_-_44174330 | 0.55 |
ENST00000340093.3 |
PLAUR |
plasminogen activator, urokinase receptor |
| chr18_+_56530794 | 0.54 |
ENST00000590285.1 ENST00000586085.1 ENST00000589288.1 |
ZNF532 |
zinc finger protein 532 |
| chrX_+_46433193 | 0.54 |
ENST00000276055.3 |
CHST7 |
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 7 |
| chr4_+_6271558 | 0.53 |
ENST00000503569.1 ENST00000226760.1 |
WFS1 |
Wolfram syndrome 1 (wolframin) |
| chr12_-_56122426 | 0.53 |
ENST00000551173.1 |
CD63 |
CD63 molecule |
| chr9_+_132815985 | 0.52 |
ENST00000372410.3 |
GPR107 |
G protein-coupled receptor 107 |
| chrX_+_51486481 | 0.52 |
ENST00000340438.4 |
GSPT2 |
G1 to S phase transition 2 |
| chr14_-_23288930 | 0.52 |
ENST00000554517.1 ENST00000285850.7 ENST00000397529.2 ENST00000555702.1 |
SLC7A7 |
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr4_-_41216492 | 0.52 |
ENST00000503503.1 ENST00000509446.1 ENST00000503264.1 ENST00000508707.1 ENST00000508593.1 |
APBB2 |
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chrX_+_80457442 | 0.51 |
ENST00000373212.5 |
SH3BGRL |
SH3 domain binding glutamic acid-rich protein like |
| chr22_+_50312379 | 0.51 |
ENST00000407217.3 ENST00000403427.3 |
CRELD2 |
cysteine-rich with EGF-like domains 2 |
| chr6_-_111804393 | 0.50 |
ENST00000368802.3 ENST00000368805.1 |
REV3L |
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr3_+_49059038 | 0.50 |
ENST00000451378.2 |
NDUFAF3 |
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr6_+_63921399 | 0.50 |
ENST00000356170.3 |
FKBP1C |
FK506 binding protein 1C |
| chr3_-_50360192 | 0.50 |
ENST00000442581.1 ENST00000447092.1 ENST00000357750.4 |
HYAL2 |
hyaluronoglucosaminidase 2 |
| chr7_-_30029367 | 0.50 |
ENST00000242059.5 |
SCRN1 |
secernin 1 |
| chr17_+_42429493 | 0.50 |
ENST00000586242.1 |
GRN |
granulin |
| chr11_-_414948 | 0.50 |
ENST00000530494.1 ENST00000528209.1 ENST00000431843.2 ENST00000528058.1 |
SIGIRR |
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr21_+_30502806 | 0.49 |
ENST00000399928.1 ENST00000399926.1 |
MAP3K7CL |
MAP3K7 C-terminal like |
| chr15_+_99645277 | 0.49 |
ENST00000336292.6 ENST00000328642.7 |
SYNM |
synemin, intermediate filament protein |
| chr14_-_92414055 | 0.49 |
ENST00000342058.4 |
FBLN5 |
fibulin 5 |
| chr4_-_139163491 | 0.49 |
ENST00000280612.5 |
SLC7A11 |
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
| chr9_-_129885010 | 0.49 |
ENST00000373425.3 |
ANGPTL2 |
angiopoietin-like 2 |
| chr7_-_30029574 | 0.49 |
ENST00000426154.1 ENST00000421434.1 ENST00000434476.2 |
SCRN1 |
secernin 1 |
| chr4_-_41216619 | 0.48 |
ENST00000508676.1 ENST00000506352.1 ENST00000295974.8 |
APBB2 |
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr4_-_77134742 | 0.48 |
ENST00000452464.2 |
SCARB2 |
scavenger receptor class B, member 2 |
| chr3_-_73673991 | 0.48 |
ENST00000308537.4 ENST00000263666.4 |
PDZRN3 |
PDZ domain containing ring finger 3 |
| chr22_+_50312274 | 0.48 |
ENST00000404488.3 |
CRELD2 |
cysteine-rich with EGF-like domains 2 |
| chr4_-_41216473 | 0.48 |
ENST00000513140.1 |
APBB2 |
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr12_-_56120865 | 0.47 |
ENST00000548898.1 ENST00000552067.1 |
CD63 |
CD63 molecule |
| chr11_-_65325664 | 0.47 |
ENST00000301873.5 |
LTBP3 |
latent transforming growth factor beta binding protein 3 |
| chr14_-_103523745 | 0.47 |
ENST00000361246.2 |
CDC42BPB |
CDC42 binding protein kinase beta (DMPK-like) |
| chr10_+_120967072 | 0.46 |
ENST00000392870.2 |
GRK5 |
G protein-coupled receptor kinase 5 |
| chr22_+_50312316 | 0.46 |
ENST00000328268.4 |
CRELD2 |
cysteine-rich with EGF-like domains 2 |
| chr22_-_22292934 | 0.45 |
ENST00000538191.1 ENST00000424647.1 ENST00000407142.1 |
PPM1F |
protein phosphatase, Mg2+/Mn2+ dependent, 1F |
| chr20_+_44519948 | 0.45 |
ENST00000354880.5 ENST00000191018.5 |
CTSA |
cathepsin A |
| chrX_+_23685653 | 0.45 |
ENST00000379331.3 |
PRDX4 |
peroxiredoxin 4 |
| chr2_+_173292280 | 0.45 |
ENST00000264107.7 |
ITGA6 |
integrin, alpha 6 |
| chr1_-_231114542 | 0.45 |
ENST00000522821.1 ENST00000366661.4 ENST00000366662.4 ENST00000414259.1 ENST00000522399.1 |
TTC13 |
tetratricopeptide repeat domain 13 |
| chr10_-_15413035 | 0.45 |
ENST00000378116.4 ENST00000455654.1 |
FAM171A1 |
family with sequence similarity 171, member A1 |
| chr16_-_66785497 | 0.45 |
ENST00000440564.2 ENST00000379482.2 ENST00000443351.2 ENST00000566150.1 |
DYNC1LI2 |
dynein, cytoplasmic 1, light intermediate chain 2 |
| chr6_-_139695757 | 0.45 |
ENST00000367651.2 |
CITED2 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
| chr9_-_129884902 | 0.45 |
ENST00000373417.1 |
ANGPTL2 |
angiopoietin-like 2 |
| chr9_-_117880477 | 0.44 |
ENST00000534839.1 ENST00000340094.3 ENST00000535648.1 ENST00000346706.3 ENST00000345230.3 ENST00000350763.4 |
TNC |
tenascin C |
| chr12_-_56120838 | 0.43 |
ENST00000548160.1 |
CD63 |
CD63 molecule |
| chr12_-_104443890 | 0.43 |
ENST00000547583.1 ENST00000360814.4 ENST00000546851.1 |
GLT8D2 |
glycosyltransferase 8 domain containing 2 |
| chr12_-_56122220 | 0.43 |
ENST00000552692.1 |
CD63 |
CD63 molecule |
| chr4_-_77135046 | 0.43 |
ENST00000264896.2 |
SCARB2 |
scavenger receptor class B, member 2 |
| chr10_-_48438974 | 0.43 |
ENST00000224605.2 |
GDF10 |
growth differentiation factor 10 |
| chr7_+_134331550 | 0.43 |
ENST00000344924.3 ENST00000418040.1 ENST00000393132.2 |
BPGM |
2,3-bisphosphoglycerate mutase |
| chr9_-_128003606 | 0.43 |
ENST00000324460.6 |
HSPA5 |
heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa) |
| chr5_+_72921983 | 0.43 |
ENST00000296794.6 ENST00000545377.1 ENST00000513042.2 ENST00000287898.5 ENST00000509848.1 |
ARHGEF28 |
Rho guanine nucleotide exchange factor (GEF) 28 |
| chr1_-_113249734 | 0.43 |
ENST00000484054.3 ENST00000369636.2 ENST00000369637.1 ENST00000285735.2 ENST00000369638.2 |
RHOC |
ras homolog family member C |
| chr7_-_100860851 | 0.42 |
ENST00000223127.3 |
PLOD3 |
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr12_-_7261772 | 0.42 |
ENST00000545280.1 ENST00000543933.1 ENST00000545337.1 ENST00000544702.1 ENST00000266542.4 |
C1RL |
complement component 1, r subcomponent-like |
| chr2_+_189156721 | 0.42 |
ENST00000409927.1 ENST00000409805.1 |
GULP1 |
GULP, engulfment adaptor PTB domain containing 1 |
| chr7_+_100210133 | 0.42 |
ENST00000393950.2 ENST00000424091.2 |
MOSPD3 |
motile sperm domain containing 3 |
| chr4_-_129208940 | 0.42 |
ENST00000296425.5 |
PGRMC2 |
progesterone receptor membrane component 2 |
| chr9_+_82186682 | 0.42 |
ENST00000376552.2 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr11_-_65325430 | 0.42 |
ENST00000322147.4 |
LTBP3 |
latent transforming growth factor beta binding protein 3 |
| chr19_+_40877583 | 0.42 |
ENST00000596470.1 |
PLD3 |
phospholipase D family, member 3 |
| chr14_-_27066960 | 0.41 |
ENST00000539517.2 |
NOVA1 |
neuro-oncological ventral antigen 1 |
| chr1_+_145524891 | 0.41 |
ENST00000369304.3 |
ITGA10 |
integrin, alpha 10 |
| chr18_+_61637159 | 0.41 |
ENST00000397985.2 ENST00000353706.2 ENST00000542677.1 ENST00000397988.3 ENST00000448851.1 |
SERPINB8 |
serpin peptidase inhibitor, clade B (ovalbumin), member 8 |
| chr2_+_185463093 | 0.41 |
ENST00000302277.6 |
ZNF804A |
zinc finger protein 804A |
| chr8_+_15397732 | 0.40 |
ENST00000382020.4 ENST00000506802.1 ENST00000509380.1 ENST00000503731.1 |
TUSC3 |
tumor suppressor candidate 3 |
| chr20_+_44520009 | 0.40 |
ENST00000607482.1 ENST00000372459.2 |
CTSA |
cathepsin A |
| chr14_+_90863364 | 0.40 |
ENST00000447653.3 ENST00000553542.1 |
CALM1 |
calmodulin 1 (phosphorylase kinase, delta) |
| chr12_-_15114603 | 0.40 |
ENST00000228945.4 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
| chrX_-_15402498 | 0.39 |
ENST00000297904.3 |
FIGF |
c-fos induced growth factor (vascular endothelial growth factor D) |
| chr2_-_235405168 | 0.39 |
ENST00000339728.3 |
ARL4C |
ADP-ribosylation factor-like 4C |
| chr17_+_42634844 | 0.39 |
ENST00000315323.3 |
FZD2 |
frizzled family receptor 2 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 7.2 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.9 | 2.6 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.7 | 1.4 | GO:0035989 | tendon development(GO:0035989) |
| 0.7 | 2.0 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.6 | 2.5 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.6 | 1.8 | GO:0061151 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm development(GO:0048389) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) pattern specification involved in mesonephros development(GO:0061227) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in kidney development(GO:0072098) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.6 | 6.0 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.6 | 1.7 | GO:1904395 | retinal rod cell differentiation(GO:0060221) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.5 | 0.5 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.5 | 1.9 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.5 | 1.4 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) |
| 0.4 | 1.6 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.4 | 1.5 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.3 | 0.6 | GO:1904783 | positive regulation of NMDA glutamate receptor activity(GO:1904783) |
| 0.3 | 1.3 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.3 | 0.9 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.3 | 1.2 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.3 | 1.2 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.3 | 1.2 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.3 | 3.3 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.3 | 1.3 | GO:1901662 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.3 | 3.4 | GO:2000680 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.3 | 1.0 | GO:0061304 | extracellular matrix-cell signaling(GO:0035426) retinal blood vessel morphogenesis(GO:0061304) |
| 0.2 | 0.7 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.2 | 0.9 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.2 | 0.9 | GO:1905123 | regulation of endosome organization(GO:1904978) regulation of glucosylceramidase activity(GO:1905123) |
| 0.2 | 0.7 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) renal albumin absorption(GO:0097018) oligodendrocyte apoptotic process(GO:0097252) regulation of renal albumin absorption(GO:2000532) |
| 0.2 | 0.6 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.2 | 0.6 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.2 | 1.0 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.2 | 1.2 | GO:1902998 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.2 | 1.8 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.2 | 0.5 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.2 | 0.5 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.2 | 0.7 | GO:0097368 | histone H4-K20 trimethylation(GO:0034773) establishment of Sertoli cell barrier(GO:0097368) |
| 0.2 | 0.5 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.2 | 0.8 | GO:0009609 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.1 | 0.4 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.1 | 0.7 | GO:0003150 | muscular septum morphogenesis(GO:0003150) |
| 0.1 | 1.2 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 | 0.7 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.1 | 0.8 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.1 | 0.4 | GO:0021589 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.1 | 0.8 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) positive regulation of autophagosome maturation(GO:1901098) |
| 0.1 | 0.8 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.1 | 1.9 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 0.5 | GO:0019087 | transformation of host cell by virus(GO:0019087) |
| 0.1 | 1.1 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.1 | 0.8 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 1.0 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.1 | 1.2 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.1 | 1.5 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 0.3 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.1 | 0.4 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.1 | 1.7 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 0.8 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.1 | 4.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 0.7 | GO:1900020 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.1 | 1.2 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.1 | 0.3 | GO:0045210 | FasL biosynthetic process(GO:0045210) |
| 0.1 | 2.2 | GO:1901881 | positive regulation of protein depolymerization(GO:1901881) |
| 0.1 | 0.9 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.1 | 0.9 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.3 | GO:0002585 | positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen(GO:0002585) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.1 | 0.3 | GO:1903960 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) negative regulation of anion transmembrane transport(GO:1903960) |
| 0.1 | 0.2 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.1 | 0.8 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.1 | 1.2 | GO:0035878 | nail development(GO:0035878) |
| 0.1 | 0.4 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.1 | 0.6 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.1 | 0.2 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.1 | 0.7 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 0.7 | GO:0045915 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.1 | 0.5 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.1 | 0.6 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) |
| 0.1 | 0.2 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.1 | 0.5 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.1 | 0.3 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.1 | 0.5 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 | 1.3 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.1 | 0.8 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 1.7 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.1 | 0.2 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.1 | 0.6 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.1 | 0.2 | GO:1902617 | response to fluoride(GO:1902617) |
| 0.1 | 0.2 | GO:0097490 | sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) |
| 0.1 | 0.2 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.1 | 2.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.4 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 1.1 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.1 | 0.8 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 0.3 | GO:1903423 | positive regulation of synaptic vesicle endocytosis(GO:1900244) positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.1 | 0.3 | GO:0015862 | uridine transport(GO:0015862) |
| 0.1 | 0.2 | GO:0097212 | cadmium ion homeostasis(GO:0055073) lysosomal membrane organization(GO:0097212) regulation of hydrogen peroxide catabolic process(GO:2000295) negative regulation of hydrogen peroxide catabolic process(GO:2000296) regulation of oxygen metabolic process(GO:2000374) |
| 0.1 | 0.3 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 0.5 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.0 | 0.1 | GO:0042700 | luteinizing hormone signaling pathway(GO:0042700) |
| 0.0 | 0.5 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.4 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.6 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 1.0 | GO:1900120 | regulation of receptor binding(GO:1900120) |
| 0.0 | 0.2 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) positive regulation of potassium ion import(GO:1903288) |
| 0.0 | 0.7 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.1 | GO:1904502 | lipophagy(GO:0061724) regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.0 | 0.2 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.0 | 0.1 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.0 | 0.5 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:1904886 | beta-catenin destruction complex disassembly(GO:1904886) |
| 0.0 | 0.2 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.1 | GO:0043449 | olfactory learning(GO:0008355) cellular alkene metabolic process(GO:0043449) |
| 0.0 | 0.3 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.1 | GO:0097350 | neutrophil clearance(GO:0097350) |
| 0.0 | 0.4 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 1.2 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 0.0 | 0.1 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 | 0.4 | GO:0006911 | phagocytosis, engulfment(GO:0006911) membrane invagination(GO:0010324) |
| 0.0 | 0.5 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.2 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.0 | 0.1 | GO:1990822 | basic amino acid transmembrane transport(GO:1990822) |
| 0.0 | 0.2 | GO:0050893 | sensory processing(GO:0050893) |
| 0.0 | 0.5 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.3 | GO:0002759 | regulation of antimicrobial humoral response(GO:0002759) |
| 0.0 | 0.1 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.0 | 0.2 | GO:0038170 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.0 | 0.0 | GO:1900369 | regulation of RNA interference(GO:1900368) negative regulation of RNA interference(GO:1900369) |
| 0.0 | 8.4 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.3 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.1 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.0 | 0.1 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.0 | 0.2 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.7 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.1 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.6 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.1 | GO:1900138 | negative regulation of icosanoid secretion(GO:0032304) negative regulation of phospholipase A2 activity(GO:1900138) |
| 0.0 | 0.3 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.2 | GO:0034770 | histone H4-K20 methylation(GO:0034770) |
| 0.0 | 0.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.1 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.5 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 1.2 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.0 | 0.1 | GO:2000870 | regulation of progesterone secretion(GO:2000870) |
| 0.0 | 0.1 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.0 | 0.1 | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 1.4 | GO:0042116 | macrophage activation(GO:0042116) |
| 0.0 | 0.2 | GO:0042659 | regulation of cell fate specification(GO:0042659) |
| 0.0 | 0.3 | GO:0060004 | reflex(GO:0060004) |
| 0.0 | 0.4 | GO:0010002 | cardioblast differentiation(GO:0010002) |
| 0.0 | 1.2 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.1 | GO:0051710 | regulation of cytolysis in other organism(GO:0051710) |
| 0.0 | 0.5 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.0 | 0.4 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.1 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.3 | GO:0000717 | nucleotide-excision repair, DNA duplex unwinding(GO:0000717) |
| 0.0 | 0.6 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 0.4 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.0 | 0.3 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.0 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.3 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.2 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.1 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 0.4 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.5 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.7 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.2 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 1.4 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.3 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.0 | GO:0061110 | dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.1 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 0.0 | 0.3 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.0 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.0 | GO:0097210 | cellular response to cocaine(GO:0071314) response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.1 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.0 | 0.1 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.0 | 0.2 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.5 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.0 | 0.1 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.1 | GO:0045914 | cellular response to phosphate starvation(GO:0016036) positive regulation of sulfur amino acid metabolic process(GO:0031337) negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) positive regulation of homocysteine metabolic process(GO:0050668) |
| 0.0 | 0.6 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.8 | GO:1903318 | negative regulation of protein processing(GO:0010955) negative regulation of protein maturation(GO:1903318) |
| 0.0 | 0.4 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.0 | 0.7 | GO:0007517 | muscle organ development(GO:0007517) |
| 0.0 | 0.3 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.0 | GO:0033685 | negative regulation of luteinizing hormone secretion(GO:0033685) |
| 0.0 | 0.0 | GO:0022614 | apical constriction(GO:0003383) membrane to membrane docking(GO:0022614) negative regulation of myosin-light-chain-phosphatase activity(GO:0035509) negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.1 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.2 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.0 | 0.4 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.0 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.0 | 0.2 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.1 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.3 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.2 | GO:0032785 | negative regulation of DNA-templated transcription, elongation(GO:0032785) |
| 0.0 | 0.3 | GO:0006266 | DNA ligation(GO:0006266) |
| 0.0 | 0.1 | GO:0070236 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.3 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.3 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.2 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.7 | 3.0 | GO:0008112 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.7 | 9.3 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.7 | 2.0 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.6 | 1.9 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.6 | 1.9 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.4 | 1.3 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.4 | 1.7 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.4 | 1.7 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) thiol oxidase activity(GO:0016972) |
| 0.4 | 1.2 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.3 | 3.7 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.3 | 1.6 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.3 | 1.0 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.2 | 2.6 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 0.7 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.2 | 1.8 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.2 | 0.6 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.2 | 0.6 | GO:0050135 | NAD+ nucleosidase activity(GO:0003953) NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.2 | 0.6 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.2 | 1.8 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.2 | 2.4 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.2 | 0.7 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.2 | 0.6 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.2 | 0.5 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.1 | 0.9 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 1.9 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.1 | 0.5 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.1 | 1.2 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.1 | 1.3 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 1.0 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 0.5 | GO:0033906 | protein tyrosine kinase inhibitor activity(GO:0030292) hyaluronoglucuronidase activity(GO:0033906) |
| 0.1 | 2.2 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.1 | 1.2 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 0.8 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.1 | 1.1 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.9 | GO:0016997 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.1 | 0.4 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.1 | 0.4 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 0.5 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.1 | 0.4 | GO:0030305 | beta-glucuronidase activity(GO:0004566) heparanase activity(GO:0030305) |
| 0.1 | 0.5 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.1 | 1.3 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 0.5 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 2.5 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 1.7 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 0.4 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 1.5 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.8 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 0.2 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 0.1 | 0.2 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.1 | 0.3 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.1 | 1.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.4 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 1.0 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.3 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.1 | 1.1 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 0.6 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.1 | 1.4 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 0.3 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.2 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 1.6 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 1.3 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.7 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.5 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.1 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.0 | 1.0 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.9 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.7 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.2 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.0 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 1.8 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.1 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.0 | 0.2 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.0 | 0.4 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 1.1 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 1.9 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.1 | GO:0001224 | RNA polymerase II transcription cofactor binding(GO:0001224) |
| 0.0 | 0.1 | GO:0017050 | lipid kinase activity(GO:0001727) sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 1.3 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.5 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0004960 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.0 | 0.6 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.8 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.2 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.2 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 1.0 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.8 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.5 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.8 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.1 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.0 | 0.4 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.1 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.6 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.8 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.1 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.2 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.0 | 0.6 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.0 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.7 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 0.7 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.0 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.3 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.1 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.3 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.0 | 0.1 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.0 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.4 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.9 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.0 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.0 | 0.4 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.2 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.0 | 0.4 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.2 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 1.5 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.0 | GO:0003968 | RNA-directed RNA polymerase activity(GO:0003968) |
| 0.0 | 0.0 | GO:0033265 | choline binding(GO:0033265) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 8.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.0 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 5.3 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 0.8 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 1.5 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 1.9 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 1.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 0.6 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 2.0 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.0 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 1.1 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 1.5 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 1.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.7 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 3.0 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.9 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 1.2 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.8 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.9 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.0 | 0.6 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 1.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.5 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.8 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 1.0 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 1.2 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 2.6 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.7 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.7 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.5 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.8 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.4 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.4 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 1.5 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.5 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.2 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 10.5 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.1 | 4.8 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 0.3 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.1 | 1.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 2.0 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 8.7 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 2.9 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 8.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.2 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.0 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 1.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.5 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.9 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.6 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.7 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 5.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.6 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.7 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.9 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.2 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.2 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 1.0 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.2 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.6 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.8 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.4 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 1.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.0 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.4 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.6 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.4 | ST GA13 PATHWAY | G alpha 13 Pathway |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.4 | 1.1 | GO:0034676 | integrin alpha6-beta4 complex(GO:0034676) |
| 0.4 | 2.8 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.3 | 1.2 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.3 | 3.4 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.2 | 0.5 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.2 | 0.5 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.1 | 1.3 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 0.4 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.1 | 0.9 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.1 | 0.7 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 0.8 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 0.5 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.1 | 0.5 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.1 | 0.2 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.1 | 0.8 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 1.2 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 0.7 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 1.2 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 0.8 | GO:0097413 | Lewy body(GO:0097413) |
| 0.1 | 0.3 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.1 | 7.1 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.1 | 1.5 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 0.3 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.1 | 3.0 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 2.1 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 1.0 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.3 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 0.6 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 1.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 0.7 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.8 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 1.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 0.3 | GO:0001652 | granular component(GO:0001652) |
| 0.1 | 0.3 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.1 | 0.3 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 0.5 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.3 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 4.9 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.3 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 0.2 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.0 | 4.4 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.6 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.6 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.6 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.5 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.2 | GO:0005929 | cilium(GO:0005929) |
| 0.0 | 6.8 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.2 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.0 | 1.7 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 0.9 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.2 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 2.6 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.3 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.0 | 2.6 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 1.3 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.5 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.1 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.3 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.6 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 1.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.7 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.5 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 2.8 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.7 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.1 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.4 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.3 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.3 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 1.6 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.9 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.3 | GO:0043034 | costamere(GO:0043034) |