Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
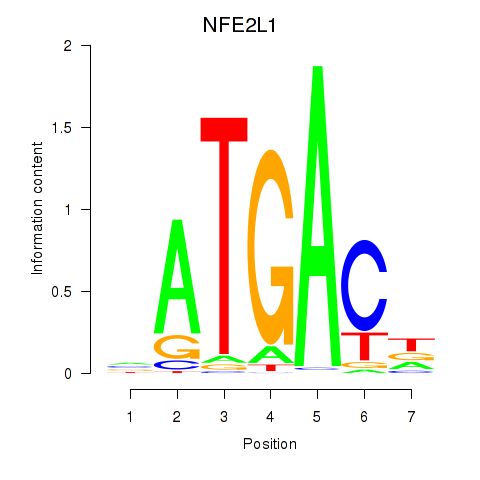
Results for NFE2L1
Z-value: 1.27
Transcription factors associated with NFE2L1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFE2L1
|
ENSG00000082641.11 | NFE2L1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFE2L1 | hg19_v2_chr17_+_46126135_46126152 | 0.18 | 5.0e-01 | Click! |
Activity profile of NFE2L1 motif
Sorted Z-values of NFE2L1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NFE2L1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_52887034 | 6.88 |
ENST00000330722.6 |
KRT6A |
keratin 6A |
| chr11_+_35201826 | 3.10 |
ENST00000531873.1 |
CD44 |
CD44 molecule (Indian blood group) |
| chr11_+_35211511 | 3.08 |
ENST00000524922.1 |
CD44 |
CD44 molecule (Indian blood group) |
| chr2_-_113594279 | 3.04 |
ENST00000416750.1 ENST00000418817.1 ENST00000263341.2 |
IL1B |
interleukin 1, beta |
| chr11_+_35198243 | 2.92 |
ENST00000528455.1 |
CD44 |
CD44 molecule (Indian blood group) |
| chr11_+_35198118 | 2.55 |
ENST00000525211.1 ENST00000526000.1 ENST00000279452.6 ENST00000527889.1 |
CD44 |
CD44 molecule (Indian blood group) |
| chr12_-_15103621 | 2.32 |
ENST00000536592.1 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
| chr11_+_35211429 | 2.30 |
ENST00000525688.1 ENST00000278385.6 ENST00000533222.1 |
CD44 |
CD44 molecule (Indian blood group) |
| chr2_+_234602305 | 2.28 |
ENST00000406651.1 |
UGT1A6 |
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr11_-_125366089 | 2.20 |
ENST00000366139.3 ENST00000278919.3 |
FEZ1 |
fasciculation and elongation protein zeta 1 (zygin I) |
| chr12_-_15104040 | 2.15 |
ENST00000541644.1 ENST00000545895.1 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
| chr9_-_117853297 | 2.14 |
ENST00000542877.1 ENST00000537320.1 ENST00000341037.4 |
TNC |
tenascin C |
| chr11_-_87908600 | 2.10 |
ENST00000531138.1 ENST00000526372.1 ENST00000243662.6 |
RAB38 |
RAB38, member RAS oncogene family |
| chr2_+_234545092 | 1.93 |
ENST00000344644.5 |
UGT1A10 |
UDP glucuronosyltransferase 1 family, polypeptide A10 |
| chr3_-_127541194 | 1.87 |
ENST00000453507.2 |
MGLL |
monoglyceride lipase |
| chr2_-_220119280 | 1.80 |
ENST00000392088.2 |
TUBA4A |
tubulin, alpha 4a |
| chr11_-_119999611 | 1.71 |
ENST00000529044.1 |
TRIM29 |
tripartite motif containing 29 |
| chr11_-_119999539 | 1.67 |
ENST00000541857.1 |
TRIM29 |
tripartite motif containing 29 |
| chr1_+_35220613 | 1.51 |
ENST00000338513.1 |
GJB5 |
gap junction protein, beta 5, 31.1kDa |
| chr6_-_56507586 | 1.48 |
ENST00000439203.1 ENST00000518935.1 ENST00000446842.2 ENST00000370765.6 ENST00000244364.6 |
DST |
dystonin |
| chr11_-_119991589 | 1.42 |
ENST00000526881.1 |
TRIM29 |
tripartite motif containing 29 |
| chr2_+_234590556 | 1.41 |
ENST00000373426.3 |
UGT1A7 |
UDP glucuronosyltransferase 1 family, polypeptide A7 |
| chr22_+_38142235 | 1.34 |
ENST00000407319.2 ENST00000403663.2 ENST00000428075.1 |
TRIOBP |
TRIO and F-actin binding protein |
| chrX_+_107069063 | 1.34 |
ENST00000262843.6 |
MID2 |
midline 2 |
| chr15_+_71228826 | 1.30 |
ENST00000558456.1 ENST00000560158.2 ENST00000558808.1 ENST00000559806.1 ENST00000559069.1 |
LRRC49 |
leucine rich repeat containing 49 |
| chr10_+_5566916 | 1.30 |
ENST00000315238.1 |
CALML3 |
calmodulin-like 3 |
| chr2_+_234580499 | 1.27 |
ENST00000354728.4 |
UGT1A9 |
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr1_-_94312706 | 1.25 |
ENST00000370244.1 |
BCAR3 |
breast cancer anti-estrogen resistance 3 |
| chr2_+_234580525 | 1.25 |
ENST00000609637.1 |
UGT1A1 |
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr1_+_26605618 | 1.23 |
ENST00000270792.5 |
SH3BGRL3 |
SH3 domain binding glutamic acid-rich protein like 3 |
| chr12_-_106641728 | 1.20 |
ENST00000378026.4 |
CKAP4 |
cytoskeleton-associated protein 4 |
| chr19_+_35645618 | 1.16 |
ENST00000392218.2 ENST00000543307.1 ENST00000392219.2 ENST00000541435.2 ENST00000590686.1 ENST00000342879.3 ENST00000588699.1 |
FXYD5 |
FXYD domain containing ion transport regulator 5 |
| chr16_+_31044413 | 1.15 |
ENST00000394998.1 |
STX4 |
syntaxin 4 |
| chr6_-_134639180 | 1.14 |
ENST00000367858.5 |
SGK1 |
serum/glucocorticoid regulated kinase 1 |
| chr19_-_44160768 | 1.12 |
ENST00000593447.1 |
PLAUR |
plasminogen activator, urokinase receptor |
| chr5_+_36608422 | 1.09 |
ENST00000381918.3 |
SLC1A3 |
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr10_-_123357598 | 1.05 |
ENST00000358487.5 ENST00000369058.3 ENST00000369060.4 ENST00000359354.2 |
FGFR2 |
fibroblast growth factor receptor 2 |
| chr2_-_161056762 | 0.98 |
ENST00000428609.2 ENST00000409967.2 |
ITGB6 |
integrin, beta 6 |
| chr12_-_123201337 | 0.96 |
ENST00000528880.2 |
HCAR3 |
hydroxycarboxylic acid receptor 3 |
| chr8_+_104831472 | 0.95 |
ENST00000262231.10 ENST00000507740.1 |
RIMS2 |
regulating synaptic membrane exocytosis 2 |
| chr2_+_219110149 | 0.94 |
ENST00000456575.1 |
ARPC2 |
actin related protein 2/3 complex, subunit 2, 34kDa |
| chrX_+_135278908 | 0.91 |
ENST00000539015.1 ENST00000370683.1 |
FHL1 |
four and a half LIM domains 1 |
| chr2_-_161056802 | 0.91 |
ENST00000283249.2 ENST00000409872.1 |
ITGB6 |
integrin, beta 6 |
| chr11_+_12399071 | 0.90 |
ENST00000539723.1 ENST00000550549.1 |
PARVA |
parvin, alpha |
| chr3_-_56809685 | 0.88 |
ENST00000413728.2 |
ARHGEF3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
| chr6_-_32784687 | 0.86 |
ENST00000447394.1 ENST00000438763.2 |
HLA-DOB |
major histocompatibility complex, class II, DO beta |
| chr11_+_123986069 | 0.86 |
ENST00000456829.2 ENST00000361352.5 ENST00000449321.1 ENST00000392748.1 ENST00000360334.4 ENST00000392744.4 |
VWA5A |
von Willebrand factor A domain containing 5A |
| chr15_-_81616446 | 0.85 |
ENST00000302824.6 |
STARD5 |
StAR-related lipid transfer (START) domain containing 5 |
| chr19_-_51530916 | 0.85 |
ENST00000594768.1 |
KLK11 |
kallikrein-related peptidase 11 |
| chr2_-_70781087 | 0.82 |
ENST00000394241.3 ENST00000295400.6 |
TGFA |
transforming growth factor, alpha |
| chr5_-_16509101 | 0.80 |
ENST00000399793.2 |
FAM134B |
family with sequence similarity 134, member B |
| chr15_+_89182178 | 0.79 |
ENST00000559876.1 |
ISG20 |
interferon stimulated exonuclease gene 20kDa |
| chr11_-_107729887 | 0.79 |
ENST00000525815.1 |
SLC35F2 |
solute carrier family 35, member F2 |
| chr2_+_234526272 | 0.79 |
ENST00000373450.4 |
UGT1A1 |
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr9_-_14180778 | 0.75 |
ENST00000380924.1 ENST00000543693.1 |
NFIB |
nuclear factor I/B |
| chr1_-_153113927 | 0.74 |
ENST00000368752.4 |
SPRR2B |
small proline-rich protein 2B |
| chr6_+_89790459 | 0.73 |
ENST00000369472.1 |
PNRC1 |
proline-rich nuclear receptor coactivator 1 |
| chrX_+_9754461 | 0.72 |
ENST00000380913.3 |
SHROOM2 |
shroom family member 2 |
| chrX_+_135279179 | 0.70 |
ENST00000370676.3 |
FHL1 |
four and a half LIM domains 1 |
| chr16_+_31044812 | 0.70 |
ENST00000313843.3 |
STX4 |
syntaxin 4 |
| chr7_-_41742697 | 0.70 |
ENST00000242208.4 |
INHBA |
inhibin, beta A |
| chr12_-_44200146 | 0.69 |
ENST00000395510.2 ENST00000325127.4 |
TWF1 |
twinfilin actin-binding protein 1 |
| chr10_+_13142225 | 0.68 |
ENST00000378747.3 |
OPTN |
optineurin |
| chr2_-_158345462 | 0.68 |
ENST00000439355.1 ENST00000540637.1 |
CYTIP |
cytohesin 1 interacting protein |
| chr12_-_123187890 | 0.67 |
ENST00000328880.5 |
HCAR2 |
hydroxycarboxylic acid receptor 2 |
| chr14_+_64680854 | 0.64 |
ENST00000458046.2 |
SYNE2 |
spectrin repeat containing, nuclear envelope 2 |
| chr2_+_30369807 | 0.63 |
ENST00000379520.3 ENST00000379519.3 ENST00000261353.4 |
YPEL5 |
yippee-like 5 (Drosophila) |
| chr13_-_99959641 | 0.63 |
ENST00000376414.4 |
GPR183 |
G protein-coupled receptor 183 |
| chr2_+_234545148 | 0.62 |
ENST00000373445.1 |
UGT1A10 |
UDP glucuronosyltransferase 1 family, polypeptide A10 |
| chr15_+_89181974 | 0.61 |
ENST00000306072.5 |
ISG20 |
interferon stimulated exonuclease gene 20kDa |
| chr6_+_63921351 | 0.61 |
ENST00000370659.1 |
FKBP1C |
FK506 binding protein 1C |
| chr10_+_13142075 | 0.61 |
ENST00000378757.2 ENST00000430081.1 ENST00000378752.3 ENST00000378748.3 |
OPTN |
optineurin |
| chr16_+_3068393 | 0.60 |
ENST00000573001.1 |
TNFRSF12A |
tumor necrosis factor receptor superfamily, member 12A |
| chr12_+_58138800 | 0.57 |
ENST00000547992.1 ENST00000552816.1 ENST00000547472.1 |
TSPAN31 |
tetraspanin 31 |
| chr1_-_170043709 | 0.57 |
ENST00000367767.1 ENST00000361580.2 ENST00000538366.1 |
KIFAP3 |
kinesin-associated protein 3 |
| chr14_-_24768913 | 0.57 |
ENST00000288111.7 |
DHRS1 |
dehydrogenase/reductase (SDR family) member 1 |
| chr16_+_57662138 | 0.56 |
ENST00000562414.1 ENST00000561969.1 ENST00000562631.1 ENST00000563445.1 ENST00000565338.1 ENST00000567702.1 |
GPR56 |
G protein-coupled receptor 56 |
| chr11_+_101983176 | 0.56 |
ENST00000524575.1 |
YAP1 |
Yes-associated protein 1 |
| chr12_+_58138664 | 0.56 |
ENST00000257910.3 |
TSPAN31 |
tetraspanin 31 |
| chr6_+_89790490 | 0.53 |
ENST00000336032.3 |
PNRC1 |
proline-rich nuclear receptor coactivator 1 |
| chr12_-_54813229 | 0.53 |
ENST00000293379.4 |
ITGA5 |
integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
| chr2_+_30369859 | 0.52 |
ENST00000402003.3 |
YPEL5 |
yippee-like 5 (Drosophila) |
| chr11_-_64052111 | 0.50 |
ENST00000394532.3 ENST00000394531.3 ENST00000309032.3 |
BAD |
BCL2-associated agonist of cell death |
| chr9_+_140083099 | 0.50 |
ENST00000322310.5 |
SSNA1 |
Sjogren syndrome nuclear autoantigen 1 |
| chr16_+_57662419 | 0.50 |
ENST00000388812.4 ENST00000538815.1 ENST00000456916.1 ENST00000567154.1 ENST00000388813.5 ENST00000562558.1 ENST00000566271.2 |
GPR56 |
G protein-coupled receptor 56 |
| chr5_+_140864649 | 0.49 |
ENST00000306593.1 |
PCDHGC4 |
protocadherin gamma subfamily C, 4 |
| chr19_+_18699535 | 0.49 |
ENST00000358607.6 |
C19orf60 |
chromosome 19 open reading frame 60 |
| chr15_+_89182156 | 0.49 |
ENST00000379224.5 |
ISG20 |
interferon stimulated exonuclease gene 20kDa |
| chrX_-_13835147 | 0.48 |
ENST00000493677.1 ENST00000355135.2 |
GPM6B |
glycoprotein M6B |
| chr9_-_132515302 | 0.48 |
ENST00000340607.4 |
PTGES |
prostaglandin E synthase |
| chr17_+_73663402 | 0.46 |
ENST00000355423.3 |
SAP30BP |
SAP30 binding protein |
| chr18_+_34124507 | 0.46 |
ENST00000591635.1 |
FHOD3 |
formin homology 2 domain containing 3 |
| chr19_-_14606900 | 0.45 |
ENST00000393029.3 ENST00000393028.1 ENST00000393033.4 ENST00000345425.2 ENST00000586027.1 ENST00000591349.1 ENST00000587210.1 |
GIPC1 |
GIPC PDZ domain containing family, member 1 |
| chr19_+_44100632 | 0.45 |
ENST00000533118.1 |
ZNF576 |
zinc finger protein 576 |
| chr1_-_6445809 | 0.45 |
ENST00000377855.2 |
ACOT7 |
acyl-CoA thioesterase 7 |
| chr2_-_106015527 | 0.44 |
ENST00000344213.4 ENST00000358129.4 |
FHL2 |
four and a half LIM domains 2 |
| chr17_+_26662730 | 0.44 |
ENST00000226225.2 |
TNFAIP1 |
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr17_+_73663470 | 0.44 |
ENST00000583536.1 |
SAP30BP |
SAP30 binding protein |
| chr8_+_104831554 | 0.44 |
ENST00000408894.2 |
RIMS2 |
regulating synaptic membrane exocytosis 2 |
| chr19_+_18699599 | 0.44 |
ENST00000450195.2 |
C19orf60 |
chromosome 19 open reading frame 60 |
| chr3_+_130650738 | 0.43 |
ENST00000504612.1 |
ATP2C1 |
ATPase, Ca++ transporting, type 2C, member 1 |
| chr5_+_148737562 | 0.43 |
ENST00000274569.4 |
PCYOX1L |
prenylcysteine oxidase 1 like |
| chr9_-_123676827 | 0.42 |
ENST00000546084.1 |
TRAF1 |
TNF receptor-associated factor 1 |
| chrX_-_13835461 | 0.42 |
ENST00000316715.4 ENST00000356942.5 |
GPM6B |
glycoprotein M6B |
| chr6_+_33043703 | 0.41 |
ENST00000418931.2 ENST00000535465.1 |
HLA-DPB1 |
major histocompatibility complex, class II, DP beta 1 |
| chr17_+_26662597 | 0.41 |
ENST00000544907.2 |
TNFAIP1 |
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr15_+_63569731 | 0.39 |
ENST00000261879.5 |
APH1B |
APH1B gamma secretase subunit |
| chr2_+_113033164 | 0.39 |
ENST00000409871.1 ENST00000343936.4 |
ZC3H6 |
zinc finger CCCH-type containing 6 |
| chr6_+_63921399 | 0.39 |
ENST00000356170.3 |
FKBP1C |
FK506 binding protein 1C |
| chr7_+_107220660 | 0.38 |
ENST00000465919.1 ENST00000445771.2 ENST00000479917.1 ENST00000421217.1 ENST00000457837.1 |
BCAP29 |
B-cell receptor-associated protein 29 |
| chr8_+_22853345 | 0.38 |
ENST00000522948.1 |
RHOBTB2 |
Rho-related BTB domain containing 2 |
| chr8_+_10530155 | 0.37 |
ENST00000521818.1 |
C8orf74 |
chromosome 8 open reading frame 74 |
| chr18_+_28898052 | 0.37 |
ENST00000257192.4 |
DSG1 |
desmoglein 1 |
| chr19_+_11200038 | 0.36 |
ENST00000558518.1 ENST00000557933.1 ENST00000455727.2 ENST00000535915.1 ENST00000545707.1 ENST00000558013.1 |
LDLR |
low density lipoprotein receptor |
| chr12_+_41086297 | 0.36 |
ENST00000551295.2 |
CNTN1 |
contactin 1 |
| chr2_+_113735575 | 0.36 |
ENST00000376489.2 ENST00000259205.4 |
IL36G |
interleukin 36, gamma |
| chr7_+_107220899 | 0.35 |
ENST00000379117.2 ENST00000473124.1 |
BCAP29 |
B-cell receptor-associated protein 29 |
| chr7_+_116312411 | 0.35 |
ENST00000456159.1 ENST00000397752.3 ENST00000318493.6 |
MET |
met proto-oncogene |
| chr11_-_66103867 | 0.34 |
ENST00000424433.2 |
RIN1 |
Ras and Rab interactor 1 |
| chrX_+_47078069 | 0.34 |
ENST00000357227.4 ENST00000519758.1 ENST00000520893.1 ENST00000517426.1 |
CDK16 |
cyclin-dependent kinase 16 |
| chr6_-_33037019 | 0.34 |
ENST00000437811.1 |
HLA-DPA1 |
major histocompatibility complex, class II, DP alpha 1 |
| chr4_+_74606223 | 0.34 |
ENST00000307407.3 ENST00000401931.1 |
IL8 |
interleukin 8 |
| chrX_-_99891796 | 0.34 |
ENST00000373020.4 |
TSPAN6 |
tetraspanin 6 |
| chr3_+_113616317 | 0.33 |
ENST00000440446.2 ENST00000488680.1 |
GRAMD1C |
GRAM domain containing 1C |
| chr2_-_106015491 | 0.33 |
ENST00000408995.1 ENST00000393353.3 ENST00000322142.8 |
FHL2 |
four and a half LIM domains 2 |
| chr5_-_112630598 | 0.33 |
ENST00000302475.4 |
MCC |
mutated in colorectal cancers |
| chr19_-_50370799 | 0.31 |
ENST00000600910.1 ENST00000322344.3 ENST00000600573.1 |
PNKP |
polynucleotide kinase 3'-phosphatase |
| chr18_+_3252206 | 0.31 |
ENST00000578562.2 |
MYL12A |
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr3_-_47950745 | 0.31 |
ENST00000429422.1 |
MAP4 |
microtubule-associated protein 4 |
| chr10_-_123357910 | 0.30 |
ENST00000336553.6 ENST00000457416.2 ENST00000360144.3 ENST00000369059.1 ENST00000356226.4 ENST00000351936.6 |
FGFR2 |
fibroblast growth factor receptor 2 |
| chr1_+_84630645 | 0.30 |
ENST00000394839.2 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
| chr8_+_9953061 | 0.30 |
ENST00000522907.1 ENST00000528246.1 |
MSRA |
methionine sulfoxide reductase A |
| chr12_-_10151773 | 0.30 |
ENST00000298527.6 ENST00000348658.4 |
CLEC1B |
C-type lectin domain family 1, member B |
| chr6_+_160183492 | 0.30 |
ENST00000541436.1 |
ACAT2 |
acetyl-CoA acetyltransferase 2 |
| chr11_+_108093559 | 0.29 |
ENST00000278616.4 |
ATM |
ataxia telangiectasia mutated |
| chr2_+_54198210 | 0.29 |
ENST00000607452.1 ENST00000422521.2 |
ACYP2 |
acylphosphatase 2, muscle type |
| chr17_-_73663245 | 0.29 |
ENST00000584999.1 ENST00000317905.5 ENST00000420326.2 ENST00000340830.5 |
RECQL5 |
RecQ protein-like 5 |
| chr1_+_84630053 | 0.29 |
ENST00000394838.2 ENST00000370682.3 ENST00000432111.1 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
| chr19_+_9361606 | 0.28 |
ENST00000456448.1 |
OR7E24 |
olfactory receptor, family 7, subfamily E, member 24 |
| chr22_-_43040515 | 0.28 |
ENST00000361740.4 |
CYB5R3 |
cytochrome b5 reductase 3 |
| chr1_+_162760513 | 0.27 |
ENST00000367915.1 ENST00000367917.3 ENST00000254521.3 ENST00000367913.1 |
HSD17B7 |
hydroxysteroid (17-beta) dehydrogenase 7 |
| chr1_+_241695424 | 0.27 |
ENST00000366558.3 ENST00000366559.4 |
KMO |
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr11_+_60223312 | 0.27 |
ENST00000532491.1 ENST00000532073.1 ENST00000534668.1 ENST00000528313.1 ENST00000533306.1 |
MS4A1 |
membrane-spanning 4-domains, subfamily A, member 1 |
| chr1_-_151319710 | 0.26 |
ENST00000290524.4 ENST00000437327.1 ENST00000452513.2 ENST00000368870.2 ENST00000452671.2 |
RFX5 |
regulatory factor X, 5 (influences HLA class II expression) |
| chr18_+_3252265 | 0.26 |
ENST00000580887.1 ENST00000536605.1 |
MYL12A |
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr11_+_108093839 | 0.26 |
ENST00000452508.2 |
ATM |
ataxia telangiectasia mutated |
| chr1_+_178062855 | 0.26 |
ENST00000448150.3 |
RASAL2 |
RAS protein activator like 2 |
| chr1_-_214638146 | 0.26 |
ENST00000543945.1 |
PTPN14 |
protein tyrosine phosphatase, non-receptor type 14 |
| chr9_-_95244781 | 0.25 |
ENST00000375544.3 ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN |
asporin |
| chr11_+_60223225 | 0.25 |
ENST00000524807.1 ENST00000345732.4 |
MS4A1 |
membrane-spanning 4-domains, subfamily A, member 1 |
| chr6_-_46048116 | 0.25 |
ENST00000185206.6 |
CLIC5 |
chloride intracellular channel 5 |
| chr7_-_142232071 | 0.25 |
ENST00000390364.3 |
TRBV10-1 |
T cell receptor beta variable 10-1(gene/pseudogene) |
| chr1_-_155211017 | 0.25 |
ENST00000536770.1 ENST00000368373.3 |
GBA |
glucosidase, beta, acid |
| chr4_-_119759795 | 0.25 |
ENST00000419654.2 |
SEC24D |
SEC24 family member D |
| chr4_+_3344141 | 0.25 |
ENST00000306648.7 |
RGS12 |
regulator of G-protein signaling 12 |
| chr12_+_123237321 | 0.25 |
ENST00000280557.6 ENST00000455982.2 |
DENR |
density-regulated protein |
| chr6_-_41130914 | 0.25 |
ENST00000373113.3 ENST00000338469.3 |
TREM2 |
triggering receptor expressed on myeloid cells 2 |
| chr7_-_75677251 | 0.25 |
ENST00000431581.1 ENST00000359697.3 ENST00000451157.1 ENST00000340062.5 ENST00000360591.3 ENST00000248600.1 |
STYXL1 |
serine/threonine/tyrosine interacting-like 1 |
| chr1_+_45265897 | 0.24 |
ENST00000372201.4 |
PLK3 |
polo-like kinase 3 |
| chr9_-_123555655 | 0.24 |
ENST00000340778.5 ENST00000453291.1 ENST00000608872.1 |
FBXW2 |
F-box and WD repeat domain containing 2 |
| chr7_+_35840819 | 0.24 |
ENST00000399035.3 |
SEPT7 |
septin 7 |
| chr19_-_10764509 | 0.24 |
ENST00000591501.1 |
ILF3-AS1 |
ILF3 antisense RNA 1 (head to head) |
| chr6_-_8064567 | 0.24 |
ENST00000543936.1 ENST00000397457.2 |
BLOC1S5 |
biogenesis of lysosomal organelles complex-1, subunit 5, muted |
| chr12_+_6644443 | 0.24 |
ENST00000396858.1 |
GAPDH |
glyceraldehyde-3-phosphate dehydrogenase |
| chr1_-_53608249 | 0.23 |
ENST00000371494.4 |
SLC1A7 |
solute carrier family 1 (glutamate transporter), member 7 |
| chr5_+_140248518 | 0.23 |
ENST00000398640.2 |
PCDHA11 |
protocadherin alpha 11 |
| chr11_+_63742050 | 0.23 |
ENST00000314133.3 ENST00000535431.1 |
COX8A AP000721.4 |
cytochrome c oxidase subunit VIIIA (ubiquitous) Uncharacterized protein |
| chr17_-_27418537 | 0.23 |
ENST00000408971.2 |
TIAF1 |
TGFB1-induced anti-apoptotic factor 1 |
| chr2_+_46769798 | 0.23 |
ENST00000238738.4 |
RHOQ |
ras homolog family member Q |
| chr2_-_98280383 | 0.23 |
ENST00000289228.5 |
ACTR1B |
ARP1 actin-related protein 1 homolog B, centractin beta (yeast) |
| chr6_-_99873145 | 0.23 |
ENST00000369239.5 ENST00000438806.1 |
PNISR |
PNN-interacting serine/arginine-rich protein |
| chr4_+_75858318 | 0.22 |
ENST00000307428.7 |
PARM1 |
prostate androgen-regulated mucin-like protein 1 |
| chr3_-_11685345 | 0.22 |
ENST00000430365.2 |
VGLL4 |
vestigial like 4 (Drosophila) |
| chr9_-_128246769 | 0.22 |
ENST00000444226.1 |
MAPKAP1 |
mitogen-activated protein kinase associated protein 1 |
| chr3_+_10068095 | 0.22 |
ENST00000287647.3 ENST00000383807.1 ENST00000383806.1 ENST00000419585.1 |
FANCD2 |
Fanconi anemia, complementation group D2 |
| chr5_-_180242534 | 0.22 |
ENST00000333055.3 ENST00000513431.1 |
MGAT1 |
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr12_+_16035307 | 0.22 |
ENST00000538352.1 ENST00000025399.6 ENST00000419869.2 |
STRAP |
serine/threonine kinase receptor associated protein |
| chr9_-_140082983 | 0.22 |
ENST00000323927.2 |
ANAPC2 |
anaphase promoting complex subunit 2 |
| chr5_+_67586465 | 0.22 |
ENST00000336483.5 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr3_-_149095652 | 0.21 |
ENST00000305366.3 |
TM4SF1 |
transmembrane 4 L six family member 1 |
| chr9_-_97356075 | 0.20 |
ENST00000375337.3 |
FBP2 |
fructose-1,6-bisphosphatase 2 |
| chr19_-_39881669 | 0.20 |
ENST00000221266.7 |
PAF1 |
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae) |
| chr2_+_113763031 | 0.20 |
ENST00000259211.6 |
IL36A |
interleukin 36, alpha |
| chr17_-_26220366 | 0.20 |
ENST00000460380.2 ENST00000508862.1 ENST00000379102.3 ENST00000582441.1 |
LYRM9 RP1-66C13.4 |
LYR motif containing 9 Uncharacterized protein |
| chr7_+_141463897 | 0.19 |
ENST00000247879.2 |
TAS2R3 |
taste receptor, type 2, member 3 |
| chr20_-_43150601 | 0.19 |
ENST00000541235.1 ENST00000255175.1 ENST00000342374.4 |
SERINC3 |
serine incorporator 3 |
| chrX_+_123480194 | 0.19 |
ENST00000371139.4 |
SH2D1A |
SH2 domain containing 1A |
| chr12_+_28343365 | 0.19 |
ENST00000545336.1 |
CCDC91 |
coiled-coil domain containing 91 |
| chr1_-_155211065 | 0.19 |
ENST00000427500.3 |
GBA |
glucosidase, beta, acid |
| chr11_+_64052692 | 0.19 |
ENST00000377702.4 |
GPR137 |
G protein-coupled receptor 137 |
| chr3_-_52443799 | 0.19 |
ENST00000470173.1 ENST00000296288.5 |
BAP1 |
BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) |
| chr10_+_121410882 | 0.19 |
ENST00000369085.3 |
BAG3 |
BCL2-associated athanogene 3 |
| chr8_+_94752349 | 0.19 |
ENST00000391680.1 |
RBM12B-AS1 |
RBM12B antisense RNA 1 |
| chr9_+_37667978 | 0.19 |
ENST00000539465.1 |
FRMPD1 |
FERM and PDZ domain containing 1 |
| chr3_+_16926441 | 0.18 |
ENST00000418129.2 ENST00000396755.2 |
PLCL2 |
phospholipase C-like 2 |
| chrX_+_150345054 | 0.18 |
ENST00000218316.3 |
GPR50 |
G protein-coupled receptor 50 |
| chr9_+_70856899 | 0.18 |
ENST00000377342.5 ENST00000478048.1 |
CBWD3 |
COBW domain containing 3 |
| chr17_-_73663168 | 0.17 |
ENST00000578201.1 ENST00000423245.2 |
RECQL5 |
RecQ protein-like 5 |
| chr19_+_10764937 | 0.17 |
ENST00000449870.1 ENST00000318511.3 ENST00000420083.1 |
ILF3 |
interleukin enhancer binding factor 3, 90kDa |
| chr1_+_100436065 | 0.17 |
ENST00000370153.1 |
SLC35A3 |
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3 |
Gene Ontology Analysis
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.5 | 2.1 | GO:0035651 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 0.5 | 1.8 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.5 | 13.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.4 | 4.5 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.4 | 1.1 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.2 | 0.7 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.2 | 2.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.2 | 7.4 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 3.6 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.2 | 4.8 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.2 | 1.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 1.9 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 1.9 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 1.4 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.5 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 0.4 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.1 | 1.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.6 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.1 | 0.3 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.1 | 0.6 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 1.2 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.1 | 0.3 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) polynucleotide phosphatase activity(GO:0098518) |
| 0.1 | 0.5 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.1 | 0.2 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.1 | 0.3 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.1 | 0.4 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.1 | 0.3 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.1 | 1.9 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 0.2 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 0.2 | GO:0019828 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) aspartic-type endopeptidase inhibitor activity(GO:0019828) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.2 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.1 | 0.3 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 0.2 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 1.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 1.1 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 0.4 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.1 | 0.7 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 0.2 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.1 | 0.9 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.5 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 1.1 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 7.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.4 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.1 | GO:0004488 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.6 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0035529 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) NADH pyrophosphatase activity(GO:0035529) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.0 | 0.2 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 1.0 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.2 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.4 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.1 | GO:0032406 | MutLbeta complex binding(GO:0032406) MutSbeta complex binding(GO:0032408) |
| 0.0 | 0.2 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.4 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.5 | GO:0043422 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) protein kinase B binding(GO:0043422) |
| 0.0 | 1.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.3 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 1.0 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.2 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 2.9 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.3 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.1 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.0 | 1.7 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.4 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.2 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.0 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.0 | GO:0080130 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.4 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.6 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.2 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 17.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 3.0 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 2.1 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 4.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 2.1 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 2.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.9 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.5 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 1.2 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.6 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 1.4 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.5 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.7 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.6 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.5 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.9 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.1 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.4 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.6 | PID ILK PATHWAY | Integrin-linked kinase signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 13.6 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.2 | 4.9 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 1.5 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 1.4 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.1 | 2.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 1.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.7 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 2.3 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.5 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 1.0 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 1.1 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.8 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.5 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.9 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 3.5 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.4 | REACTOME SIGNALING BY NOTCH3 | Genes involved in Signaling by NOTCH3 |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.4 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 3.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.9 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 1.3 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.6 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.4 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.3 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 1.0 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 13.9 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.6 | 1.9 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.4 | 2.1 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.4 | 1.5 | GO:0031673 | H zone(GO:0031673) |
| 0.3 | 0.9 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.2 | 0.7 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.2 | 1.8 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.2 | 1.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 6.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.6 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.1 | 0.5 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.1 | 0.6 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.1 | 2.0 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 1.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 1.5 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 1.6 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.2 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.1 | 0.8 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 1.2 | GO:0042599 | lamellar body(GO:0042599) |
| 0.1 | 0.2 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 0.2 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 5.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.5 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.4 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.4 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.2 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.4 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.5 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 1.4 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.2 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 1.1 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.3 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 1.4 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 1.3 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.8 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.8 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.2 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.2 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 6.9 | GO:0051710 | regulation of cytolysis in other organism(GO:0051710) |
| 1.7 | 13.9 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 1.1 | 4.5 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 1.0 | 3.0 | GO:0070487 | monocyte aggregation(GO:0070487) |
| 0.7 | 9.6 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.7 | 2.1 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.6 | 1.9 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.6 | 1.8 | GO:0043311 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.6 | 1.7 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.5 | 1.4 | GO:0035603 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) fibroblast growth factor receptor signaling pathway involved in mammary gland specification(GO:0060595) mammary gland bud formation(GO:0060615) branch elongation involved in salivary gland morphogenesis(GO:0060667) mesenchymal cell differentiation involved in lung development(GO:0060915) |
| 0.4 | 2.0 | GO:0030047 | actin modification(GO:0030047) |
| 0.4 | 1.5 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.4 | 2.2 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.3 | 0.9 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.3 | 2.0 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.3 | 1.1 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.3 | 1.9 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.3 | 0.5 | GO:0042100 | B cell proliferation(GO:0042100) |
| 0.2 | 0.7 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.2 | 0.6 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.2 | 1.9 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.2 | 0.6 | GO:1904882 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.2 | 0.7 | GO:0012502 | induction of programmed cell death(GO:0012502) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) |
| 0.2 | 1.5 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.2 | 1.1 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.2 | 1.3 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.2 | 0.8 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.2 | 1.1 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.2 | 1.4 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.2 | 0.5 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 | 0.6 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.1 | 0.4 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.1 | 0.7 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.1 | 1.0 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 0.4 | GO:1903978 | regulation of microglial cell activation(GO:1903978) positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.1 | 0.6 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.1 | 0.4 | GO:0036114 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.1 | 0.4 | GO:1904457 | beta-glucoside metabolic process(GO:1901804) beta-glucoside catabolic process(GO:1901805) positive regulation of neuronal action potential(GO:1904457) |
| 0.1 | 0.8 | GO:0021740 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 | 0.6 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.1 | 1.3 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 0.6 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.1 | 0.9 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 0.4 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.1 | 0.3 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.1 | 0.1 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.1 | 0.2 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 0.2 | GO:0002580 | regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002580) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) |
| 0.1 | 0.2 | GO:2000777 | endomitotic cell cycle(GO:0007113) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.1 | 0.8 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.1 | 0.7 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.1 | GO:0030473 | nucleokinesis involved in cell motility in cerebral cortex radial glia guided migration(GO:0021817) nuclear migration along microtubule(GO:0030473) |
| 0.1 | 0.4 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.1 | 0.6 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.1 | 0.3 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.1 | 0.1 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.1 | 0.4 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 | 0.3 | GO:0098502 | DNA dephosphorylation(GO:0098502) |
| 0.1 | 0.2 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 | 0.8 | GO:0070508 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.1 | 0.2 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.1 | 0.9 | GO:0071670 | smooth muscle cell chemotaxis(GO:0071670) |
| 0.1 | 0.2 | GO:0043650 | dicarboxylic acid biosynthetic process(GO:0043650) |
| 0.1 | 0.2 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.1 | 0.2 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.1 | 0.2 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.1 | 0.2 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 0.0 | 0.2 | GO:0032675 | regulation of interleukin-6 production(GO:0032675) |
| 0.0 | 0.2 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.0 | 0.8 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 4.6 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.0 | 0.1 | GO:0043400 | cortisol secretion(GO:0043400) regulation of cortisol secretion(GO:0051462) |
| 0.0 | 0.3 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.5 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.9 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 0.2 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.0 | 0.4 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.9 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.3 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.3 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.0 | 1.7 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.0 | 0.2 | GO:0060088 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.5 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.3 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.2 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.1 | GO:0009217 | dGTP catabolic process(GO:0006203) purine deoxyribonucleoside triphosphate catabolic process(GO:0009217) |
| 0.0 | 0.2 | GO:0021859 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.0 | 0.5 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 0.0 | 0.2 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.1 | GO:1904075 | trophectodermal cell proliferation(GO:0001834) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.0 | 0.4 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 1.0 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.0 | 0.1 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.0 | 0.3 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.2 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.0 | 0.5 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.2 | GO:0050942 | positive regulation of pigment cell differentiation(GO:0050942) |
| 0.0 | 0.1 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.1 | GO:0018874 | benzoate metabolic process(GO:0018874) butyrate metabolic process(GO:0019605) |
| 0.0 | 0.1 | GO:0070170 | regulation of tooth mineralization(GO:0070170) |
| 0.0 | 0.1 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.2 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.2 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.1 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.0 | 1.2 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.3 | GO:0030220 | platelet formation(GO:0030220) |
| 0.0 | 0.1 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.1 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.1 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.0 | 0.1 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.4 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.0 | 0.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.1 | GO:0038169 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.0 | 0.1 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.2 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.0 | 0.1 | GO:0031115 | regulation of microtubule nucleation(GO:0010968) negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.1 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.0 | 0.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.2 | GO:0007249 | I-kappaB kinase/NF-kappaB signaling(GO:0007249) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 1.1 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |