Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
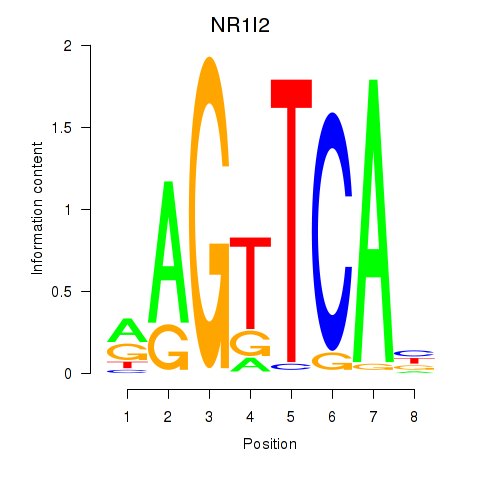
Results for NR1I2
Z-value: 1.73
Transcription factors associated with NR1I2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR1I2
|
ENSG00000144852.12 | NR1I2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR1I2 | hg19_v2_chr3_+_119501557_119501557 | -0.33 | 2.1e-01 | Click! |
Activity profile of NR1I2 motif
Sorted Z-values of NR1I2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NR1I2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_91576429 | 7.20 |
ENST00000552145.1 ENST00000546745.1 |
DCN |
decorin |
| chr11_+_117073850 | 5.87 |
ENST00000529622.1 |
TAGLN |
transgelin |
| chr3_+_157154578 | 5.43 |
ENST00000295927.3 |
PTX3 |
pentraxin 3, long |
| chr1_-_151965048 | 5.11 |
ENST00000368809.1 |
S100A10 |
S100 calcium binding protein A10 |
| chr15_+_39873268 | 5.00 |
ENST00000397591.2 ENST00000260356.5 |
THBS1 |
thrombospondin 1 |
| chrX_+_135278908 | 4.43 |
ENST00000539015.1 ENST00000370683.1 |
FHL1 |
four and a half LIM domains 1 |
| chr11_-_111783595 | 4.08 |
ENST00000528628.1 |
CRYAB |
crystallin, alpha B |
| chrX_+_135279179 | 4.02 |
ENST00000370676.3 |
FHL1 |
four and a half LIM domains 1 |
| chr5_+_125695805 | 4.01 |
ENST00000513040.1 |
GRAMD3 |
GRAM domain containing 3 |
| chr7_+_134576151 | 3.69 |
ENST00000393118.2 |
CALD1 |
caldesmon 1 |
| chr7_-_107642348 | 3.32 |
ENST00000393561.1 |
LAMB1 |
laminin, beta 1 |
| chr10_-_33625154 | 3.15 |
ENST00000265371.4 |
NRP1 |
neuropilin 1 |
| chr12_-_91576561 | 3.13 |
ENST00000547568.2 ENST00000552962.1 |
DCN |
decorin |
| chr17_-_46682321 | 3.01 |
ENST00000225648.3 ENST00000484302.2 |
HOXB6 |
homeobox B6 |
| chr2_-_86790593 | 2.89 |
ENST00000263856.4 ENST00000409225.2 |
CHMP3 |
charged multivesicular body protein 3 |
| chr12_-_91576750 | 2.73 |
ENST00000228329.5 ENST00000303320.3 ENST00000052754.5 |
DCN |
decorin |
| chr13_-_110959478 | 2.72 |
ENST00000543140.1 ENST00000375820.4 |
COL4A1 |
collagen, type IV, alpha 1 |
| chr8_-_18666360 | 2.67 |
ENST00000286485.8 |
PSD3 |
pleckstrin and Sec7 domain containing 3 |
| chr5_+_34656569 | 2.58 |
ENST00000428746.2 |
RAI14 |
retinoic acid induced 14 |
| chr15_+_96875657 | 2.54 |
ENST00000559679.1 ENST00000394171.2 |
NR2F2 |
nuclear receptor subfamily 2, group F, member 2 |
| chr2_-_158732340 | 2.50 |
ENST00000539637.1 ENST00000413751.1 ENST00000434821.1 ENST00000424669.1 |
ACVR1 |
activin A receptor, type I |
| chr11_-_111794446 | 2.48 |
ENST00000527950.1 |
CRYAB |
crystallin, alpha B |
| chr2_+_192141611 | 2.44 |
ENST00000392316.1 |
MYO1B |
myosin IB |
| chr1_-_120612240 | 2.39 |
ENST00000256646.2 |
NOTCH2 |
notch 2 |
| chr12_-_6665200 | 2.39 |
ENST00000336604.4 ENST00000396840.2 ENST00000356896.4 |
IFFO1 |
intermediate filament family orphan 1 |
| chrX_+_102840408 | 2.36 |
ENST00000468024.1 ENST00000472484.1 ENST00000415568.2 ENST00000490644.1 ENST00000459722.1 ENST00000472745.1 ENST00000494801.1 ENST00000434216.2 ENST00000425011.1 |
TCEAL4 |
transcription elongation factor A (SII)-like 4 |
| chr3_-_114343039 | 2.32 |
ENST00000481632.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr15_+_96876340 | 2.28 |
ENST00000453270.2 |
NR2F2 |
nuclear receptor subfamily 2, group F, member 2 |
| chr2_+_30454390 | 2.11 |
ENST00000395323.3 ENST00000406087.1 ENST00000404397.1 |
LBH |
limb bud and heart development |
| chr3_+_8543393 | 2.09 |
ENST00000157600.3 ENST00000415597.1 ENST00000535732.1 |
LMCD1 |
LIM and cysteine-rich domains 1 |
| chr2_-_235405168 | 2.06 |
ENST00000339728.3 |
ARL4C |
ADP-ribosylation factor-like 4C |
| chr6_-_167040731 | 1.99 |
ENST00000265678.4 |
RPS6KA2 |
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
| chr3_+_8543561 | 1.92 |
ENST00000397386.3 |
LMCD1 |
LIM and cysteine-rich domains 1 |
| chr9_-_14313641 | 1.88 |
ENST00000380953.1 |
NFIB |
nuclear factor I/B |
| chr12_-_56106060 | 1.87 |
ENST00000452168.2 |
ITGA7 |
integrin, alpha 7 |
| chr9_-_14313893 | 1.86 |
ENST00000380921.3 ENST00000380959.3 |
NFIB |
nuclear factor I/B |
| chrX_+_135251835 | 1.85 |
ENST00000456445.1 |
FHL1 |
four and a half LIM domains 1 |
| chr13_-_33859819 | 1.70 |
ENST00000336934.5 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
| chr8_-_18541603 | 1.61 |
ENST00000428502.2 |
PSD3 |
pleckstrin and Sec7 domain containing 3 |
| chr6_+_142622991 | 1.61 |
ENST00000230173.6 ENST00000367608.2 |
GPR126 |
G protein-coupled receptor 126 |
| chr9_-_129885010 | 1.60 |
ENST00000373425.3 |
ANGPTL2 |
angiopoietin-like 2 |
| chr20_+_43343886 | 1.54 |
ENST00000190983.4 |
WISP2 |
WNT1 inducible signaling pathway protein 2 |
| chr2_+_69240415 | 1.52 |
ENST00000409829.3 |
ANTXR1 |
anthrax toxin receptor 1 |
| chr7_+_128399002 | 1.52 |
ENST00000493278.1 |
CALU |
calumenin |
| chr5_+_140810132 | 1.51 |
ENST00000252085.3 |
PCDHGA12 |
protocadherin gamma subfamily A, 12 |
| chr2_+_223916862 | 1.47 |
ENST00000604125.1 |
KCNE4 |
potassium voltage-gated channel, Isk-related family, member 4 |
| chr2_+_69240302 | 1.47 |
ENST00000303714.4 |
ANTXR1 |
anthrax toxin receptor 1 |
| chr2_+_69240511 | 1.47 |
ENST00000409349.3 |
ANTXR1 |
anthrax toxin receptor 1 |
| chr9_-_129884902 | 1.45 |
ENST00000373417.1 |
ANGPTL2 |
angiopoietin-like 2 |
| chr6_+_142623063 | 1.42 |
ENST00000296932.8 ENST00000367609.3 |
GPR126 |
G protein-coupled receptor 126 |
| chr7_-_151433342 | 1.41 |
ENST00000433631.2 |
PRKAG2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr9_-_14314066 | 1.41 |
ENST00000397575.3 |
NFIB |
nuclear factor I/B |
| chr7_-_151433393 | 1.40 |
ENST00000492843.1 |
PRKAG2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr7_-_27169801 | 1.36 |
ENST00000511914.1 |
HOXA4 |
homeobox A4 |
| chr7_-_76247617 | 1.34 |
ENST00000441393.1 |
POMZP3 |
POM121 and ZP3 fusion |
| chr19_-_43690674 | 1.34 |
ENST00000342951.6 ENST00000366175.3 |
PSG5 |
pregnancy specific beta-1-glycoprotein 5 |
| chr7_-_131241361 | 1.34 |
ENST00000378555.3 ENST00000322985.9 ENST00000541194.1 ENST00000537928.1 |
PODXL |
podocalyxin-like |
| chr6_-_46293378 | 1.33 |
ENST00000330430.6 |
RCAN2 |
regulator of calcineurin 2 |
| chr7_-_94285402 | 1.31 |
ENST00000428696.2 ENST00000445866.2 |
SGCE |
sarcoglycan, epsilon |
| chr13_-_30424821 | 1.29 |
ENST00000380680.4 |
UBL3 |
ubiquitin-like 3 |
| chr4_+_30721968 | 1.26 |
ENST00000361762.2 |
PCDH7 |
protocadherin 7 |
| chr14_+_23340822 | 1.25 |
ENST00000359591.4 |
LRP10 |
low density lipoprotein receptor-related protein 10 |
| chr5_+_92919043 | 1.24 |
ENST00000327111.3 |
NR2F1 |
nuclear receptor subfamily 2, group F, member 1 |
| chr1_+_160097462 | 1.24 |
ENST00000447527.1 |
ATP1A2 |
ATPase, Na+/K+ transporting, alpha 2 polypeptide |
| chr8_-_93115445 | 1.23 |
ENST00000523629.1 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr13_+_115000556 | 1.22 |
ENST00000252458.6 |
CDC16 |
cell division cycle 16 |
| chrX_+_149531524 | 1.19 |
ENST00000370401.2 |
MAMLD1 |
mastermind-like domain containing 1 |
| chr3_+_8543533 | 1.19 |
ENST00000454244.1 |
LMCD1 |
LIM and cysteine-rich domains 1 |
| chr4_-_159094194 | 1.18 |
ENST00000592057.1 ENST00000585682.1 ENST00000393807.5 |
FAM198B |
family with sequence similarity 198, member B |
| chr6_+_139456226 | 1.18 |
ENST00000367658.2 |
HECA |
headcase homolog (Drosophila) |
| chr19_+_16187816 | 1.16 |
ENST00000588410.1 |
TPM4 |
tropomyosin 4 |
| chr13_+_115000339 | 1.12 |
ENST00000360383.3 ENST00000375312.3 |
CDC16 |
cell division cycle 16 |
| chr13_+_32605437 | 1.11 |
ENST00000380250.3 |
FRY |
furry homolog (Drosophila) |
| chr1_+_218519577 | 1.08 |
ENST00000366930.4 ENST00000366929.4 |
TGFB2 |
transforming growth factor, beta 2 |
| chr1_+_196788887 | 1.08 |
ENST00000320493.5 ENST00000367424.4 ENST00000367421.3 |
CFHR1 CFHR2 |
complement factor H-related 1 complement factor H-related 2 |
| chr3_-_126373929 | 1.07 |
ENST00000523403.1 ENST00000524230.2 |
TXNRD3 |
thioredoxin reductase 3 |
| chr4_+_124317940 | 1.00 |
ENST00000505319.1 ENST00000339241.1 |
SPRY1 |
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
| chr10_-_125851961 | 0.99 |
ENST00000346248.5 |
CHST15 |
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15 |
| chr1_+_25944341 | 0.98 |
ENST00000263979.3 |
MAN1C1 |
mannosidase, alpha, class 1C, member 1 |
| chr15_+_43809797 | 0.98 |
ENST00000399453.1 ENST00000300231.5 |
MAP1A |
microtubule-associated protein 1A |
| chr3_+_37284668 | 0.97 |
ENST00000361924.2 ENST00000444882.1 ENST00000356847.4 ENST00000450863.2 ENST00000429018.1 |
GOLGA4 |
golgin A4 |
| chr8_-_13372395 | 0.96 |
ENST00000276297.4 ENST00000511869.1 |
DLC1 |
deleted in liver cancer 1 |
| chr2_+_172543919 | 0.94 |
ENST00000452242.1 ENST00000340296.4 |
DYNC1I2 |
dynein, cytoplasmic 1, intermediate chain 2 |
| chr17_-_9479128 | 0.94 |
ENST00000574431.1 |
STX8 |
syntaxin 8 |
| chr19_+_2977444 | 0.94 |
ENST00000246112.4 ENST00000453329.1 ENST00000482627.1 ENST00000452088.1 |
TLE6 |
transducin-like enhancer of split 6 (E(sp1) homolog, Drosophila) |
| chr3_+_128968437 | 0.92 |
ENST00000314797.6 |
COPG1 |
coatomer protein complex, subunit gamma 1 |
| chr12_-_49582978 | 0.92 |
ENST00000301071.7 |
TUBA1A |
tubulin, alpha 1a |
| chr9_+_33795533 | 0.92 |
ENST00000379405.3 |
PRSS3 |
protease, serine, 3 |
| chr16_-_28937027 | 0.91 |
ENST00000358201.4 |
RABEP2 |
rabaptin, RAB GTPase binding effector protein 2 |
| chr7_+_22766766 | 0.91 |
ENST00000426291.1 ENST00000401651.1 ENST00000258743.5 ENST00000420258.2 ENST00000407492.1 ENST00000401630.3 ENST00000406575.1 |
IL6 |
interleukin 6 (interferon, beta 2) |
| chr2_+_109237717 | 0.90 |
ENST00000409441.1 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
| chr8_+_37654424 | 0.89 |
ENST00000315215.7 |
GPR124 |
G protein-coupled receptor 124 |
| chr11_+_118478313 | 0.89 |
ENST00000356063.5 |
PHLDB1 |
pleckstrin homology-like domain, family B, member 1 |
| chr1_-_144995002 | 0.86 |
ENST00000369356.4 |
PDE4DIP |
phosphodiesterase 4D interacting protein |
| chr13_+_115000384 | 0.86 |
ENST00000356221.3 ENST00000375310.1 |
CDC16 |
cell division cycle 16 |
| chr5_-_82969405 | 0.82 |
ENST00000510978.1 |
HAPLN1 |
hyaluronan and proteoglycan link protein 1 |
| chr9_-_23825956 | 0.80 |
ENST00000397312.2 |
ELAVL2 |
ELAV like neuron-specific RNA binding protein 2 |
| chr16_+_28875126 | 0.79 |
ENST00000359285.5 ENST00000538342.1 |
SH2B1 |
SH2B adaptor protein 1 |
| chr14_+_55034599 | 0.78 |
ENST00000392067.3 ENST00000357634.3 |
SAMD4A |
sterile alpha motif domain containing 4A |
| chr5_+_149569520 | 0.78 |
ENST00000230671.2 ENST00000524041.1 |
SLC6A7 |
solute carrier family 6 (neurotransmitter transporter), member 7 |
| chr3_-_114790179 | 0.78 |
ENST00000462705.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr3_+_37284824 | 0.77 |
ENST00000431105.1 |
GOLGA4 |
golgin A4 |
| chr1_-_53387386 | 0.75 |
ENST00000467988.1 ENST00000358358.5 ENST00000371522.4 |
ECHDC2 |
enoyl CoA hydratase domain containing 2 |
| chr9_-_34620440 | 0.75 |
ENST00000421919.1 ENST00000378911.3 ENST00000477738.2 ENST00000341694.2 ENST00000259632.7 ENST00000378913.2 ENST00000378916.4 ENST00000447983.2 |
DCTN3 |
dynactin 3 (p22) |
| chr13_-_33112823 | 0.75 |
ENST00000504114.1 |
N4BP2L2 |
NEDD4 binding protein 2-like 2 |
| chr3_+_12330560 | 0.73 |
ENST00000397026.2 |
PPARG |
peroxisome proliferator-activated receptor gamma |
| chr1_+_25943959 | 0.73 |
ENST00000374332.4 |
MAN1C1 |
mannosidase, alpha, class 1C, member 1 |
| chr13_+_115000521 | 0.70 |
ENST00000252457.5 ENST00000375308.1 |
CDC16 |
cell division cycle 16 |
| chr20_-_44485835 | 0.70 |
ENST00000457981.1 ENST00000426915.1 ENST00000217455.4 |
ACOT8 |
acyl-CoA thioesterase 8 |
| chr2_+_234621551 | 0.69 |
ENST00000608381.1 ENST00000373414.3 |
UGT1A1 UGT1A5 |
UDP glucuronosyltransferase 1 family, polypeptide A8 UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr8_-_120685608 | 0.68 |
ENST00000427067.2 |
ENPP2 |
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr2_-_192711968 | 0.68 |
ENST00000304141.4 |
SDPR |
serum deprivation response |
| chrX_+_49832231 | 0.67 |
ENST00000376108.3 |
CLCN5 |
chloride channel, voltage-sensitive 5 |
| chr22_-_38699003 | 0.66 |
ENST00000451964.1 |
CSNK1E |
casein kinase 1, epsilon |
| chrX_+_38420623 | 0.65 |
ENST00000378482.2 |
TSPAN7 |
tetraspanin 7 |
| chr16_+_67207838 | 0.65 |
ENST00000566871.1 ENST00000268605.7 |
NOL3 |
nucleolar protein 3 (apoptosis repressor with CARD domain) |
| chr1_-_229569834 | 0.64 |
ENST00000366684.3 ENST00000366683.2 |
ACTA1 |
actin, alpha 1, skeletal muscle |
| chrX_+_64887512 | 0.64 |
ENST00000360270.5 |
MSN |
moesin |
| chr11_+_65687158 | 0.64 |
ENST00000532933.1 |
DRAP1 |
DR1-associated protein 1 (negative cofactor 2 alpha) |
| chr2_+_234627424 | 0.63 |
ENST00000373409.3 |
UGT1A4 |
UDP glucuronosyltransferase 1 family, polypeptide A4 |
| chr12_-_16759711 | 0.62 |
ENST00000447609.1 |
LMO3 |
LIM domain only 3 (rhombotin-like 2) |
| chr9_+_139981379 | 0.62 |
ENST00000371589.4 |
MAN1B1 |
mannosidase, alpha, class 1B, member 1 |
| chr20_+_3776936 | 0.62 |
ENST00000439880.2 |
CDC25B |
cell division cycle 25B |
| chr17_+_41323204 | 0.59 |
ENST00000542611.1 ENST00000590996.1 ENST00000389312.4 ENST00000589872.1 |
NBR1 |
neighbor of BRCA1 gene 1 |
| chr11_+_308143 | 0.59 |
ENST00000399817.4 |
IFITM2 |
interferon induced transmembrane protein 2 |
| chr1_+_196912902 | 0.58 |
ENST00000476712.2 ENST00000367415.5 |
CFHR2 |
complement factor H-related 2 |
| chr4_-_149363662 | 0.58 |
ENST00000355292.3 ENST00000358102.3 |
NR3C2 |
nuclear receptor subfamily 3, group C, member 2 |
| chr1_+_236849754 | 0.56 |
ENST00000542672.1 ENST00000366578.4 |
ACTN2 |
actinin, alpha 2 |
| chr11_+_108093559 | 0.56 |
ENST00000278616.4 |
ATM |
ataxia telangiectasia mutated |
| chr1_+_201979645 | 0.56 |
ENST00000367284.5 ENST00000367283.3 |
ELF3 |
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr18_-_52989525 | 0.55 |
ENST00000457482.3 |
TCF4 |
transcription factor 4 |
| chr19_-_46285646 | 0.55 |
ENST00000458663.2 |
DMPK |
dystrophia myotonica-protein kinase |
| chr17_-_18161870 | 0.54 |
ENST00000579294.1 ENST00000545457.2 ENST00000379450.4 ENST00000578558.1 |
FLII |
flightless I homolog (Drosophila) |
| chr16_+_67207872 | 0.54 |
ENST00000563258.1 ENST00000568146.1 |
NOL3 |
nucleolar protein 3 (apoptosis repressor with CARD domain) |
| chr7_+_6414128 | 0.52 |
ENST00000348035.4 ENST00000356142.4 |
RAC1 |
ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) |
| chr7_+_110731062 | 0.51 |
ENST00000308478.5 ENST00000451085.1 ENST00000422987.3 ENST00000421101.1 |
LRRN3 |
leucine rich repeat neuronal 3 |
| chr4_+_71600144 | 0.50 |
ENST00000502653.1 |
RUFY3 |
RUN and FYVE domain containing 3 |
| chr4_-_70361579 | 0.50 |
ENST00000512583.1 |
UGT2B4 |
UDP glucuronosyltransferase 2 family, polypeptide B4 |
| chr7_-_100183742 | 0.49 |
ENST00000310300.6 |
LRCH4 |
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr12_-_114843889 | 0.48 |
ENST00000405440.2 |
TBX5 |
T-box 5 |
| chr17_-_7145106 | 0.48 |
ENST00000577035.1 |
GABARAP |
GABA(A) receptor-associated protein |
| chr9_+_115983808 | 0.48 |
ENST00000374210.6 ENST00000374212.4 |
SLC31A1 |
solute carrier family 31 (copper transporter), member 1 |
| chr11_-_64013663 | 0.48 |
ENST00000392210.2 |
PPP1R14B |
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr5_+_177027101 | 0.47 |
ENST00000029410.5 ENST00000510761.1 ENST00000505468.1 |
B4GALT7 |
xylosylprotein beta 1,4-galactosyltransferase, polypeptide 7 |
| chr9_+_17135016 | 0.47 |
ENST00000425824.1 ENST00000262360.5 ENST00000380641.4 |
CNTLN |
centlein, centrosomal protein |
| chr4_-_111119804 | 0.47 |
ENST00000394607.3 ENST00000302274.3 |
ELOVL6 |
ELOVL fatty acid elongase 6 |
| chr3_+_69985734 | 0.47 |
ENST00000314557.6 ENST00000394351.3 |
MITF |
microphthalmia-associated transcription factor |
| chr11_+_120207787 | 0.47 |
ENST00000397843.2 ENST00000356641.3 |
ARHGEF12 |
Rho guanine nucleotide exchange factor (GEF) 12 |
| chr17_-_34344991 | 0.46 |
ENST00000591423.1 |
CCL23 |
chemokine (C-C motif) ligand 23 |
| chr17_-_2169425 | 0.46 |
ENST00000570606.1 ENST00000354901.4 |
SMG6 |
SMG6 nonsense mediated mRNA decay factor |
| chr17_-_34345002 | 0.46 |
ENST00000293280.2 |
CCL23 |
chemokine (C-C motif) ligand 23 |
| chr15_-_81616446 | 0.46 |
ENST00000302824.6 |
STARD5 |
StAR-related lipid transfer (START) domain containing 5 |
| chr20_+_3776371 | 0.44 |
ENST00000245960.5 |
CDC25B |
cell division cycle 25B |
| chr11_+_120107344 | 0.44 |
ENST00000260264.4 |
POU2F3 |
POU class 2 homeobox 3 |
| chr3_-_116164306 | 0.43 |
ENST00000490035.2 |
LSAMP |
limbic system-associated membrane protein |
| chr15_+_101420028 | 0.43 |
ENST00000557963.1 ENST00000346623.6 |
ALDH1A3 |
aldehyde dehydrogenase 1 family, member A3 |
| chr4_+_5712898 | 0.43 |
ENST00000264956.6 ENST00000382674.2 |
EVC |
Ellis van Creveld syndrome |
| chr3_+_137906154 | 0.43 |
ENST00000466749.1 ENST00000358441.2 ENST00000489213.1 |
ARMC8 |
armadillo repeat containing 8 |
| chr8_-_110986918 | 0.42 |
ENST00000297404.1 |
KCNV1 |
potassium channel, subfamily V, member 1 |
| chr3_+_137906353 | 0.42 |
ENST00000461822.1 ENST00000485396.1 ENST00000471453.1 ENST00000470821.1 ENST00000471709.1 ENST00000538260.1 ENST00000393058.3 ENST00000463485.1 |
ARMC8 |
armadillo repeat containing 8 |
| chr1_-_47407111 | 0.41 |
ENST00000371904.4 |
CYP4A11 |
cytochrome P450, family 4, subfamily A, polypeptide 11 |
| chr6_-_106773491 | 0.41 |
ENST00000360666.4 |
ATG5 |
autophagy related 5 |
| chr11_+_66624527 | 0.41 |
ENST00000393952.3 |
LRFN4 |
leucine rich repeat and fibronectin type III domain containing 4 |
| chr1_+_202995611 | 0.41 |
ENST00000367240.2 |
PPFIA4 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 4 |
| chr11_+_308217 | 0.40 |
ENST00000602569.1 |
IFITM2 |
interferon induced transmembrane protein 2 |
| chr10_+_80828774 | 0.40 |
ENST00000334512.5 |
ZMIZ1 |
zinc finger, MIZ-type containing 1 |
| chr18_+_61442629 | 0.40 |
ENST00000398019.2 ENST00000540675.1 |
SERPINB7 |
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr1_+_196743943 | 0.39 |
ENST00000471440.2 ENST00000391985.3 |
CFHR3 |
complement factor H-related 3 |
| chr2_+_18059906 | 0.38 |
ENST00000304101.4 |
KCNS3 |
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3 |
| chr18_-_53253112 | 0.38 |
ENST00000568673.1 ENST00000562847.1 ENST00000568147.1 |
TCF4 |
transcription factor 4 |
| chr9_+_17134980 | 0.38 |
ENST00000380647.3 |
CNTLN |
centlein, centrosomal protein |
| chr8_-_121824374 | 0.37 |
ENST00000517992.1 |
SNTB1 |
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1) |
| chr2_+_109223595 | 0.37 |
ENST00000410093.1 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
| chr16_-_28936493 | 0.37 |
ENST00000544477.1 ENST00000357573.6 |
RABEP2 |
rabaptin, RAB GTPase binding effector protein 2 |
| chr14_-_76447494 | 0.37 |
ENST00000238682.3 |
TGFB3 |
transforming growth factor, beta 3 |
| chr5_-_73937244 | 0.37 |
ENST00000302351.4 ENST00000510316.1 ENST00000508331.1 |
ENC1 |
ectodermal-neural cortex 1 (with BTB domain) |
| chr2_-_75426183 | 0.36 |
ENST00000409848.3 |
TACR1 |
tachykinin receptor 1 |
| chr2_-_69870747 | 0.36 |
ENST00000409068.1 |
AAK1 |
AP2 associated kinase 1 |
| chr7_-_72936608 | 0.36 |
ENST00000404251.1 |
BAZ1B |
bromodomain adjacent to zinc finger domain, 1B |
| chr19_+_36236514 | 0.36 |
ENST00000222266.2 |
PSENEN |
presenilin enhancer gamma secretase subunit |
| chr15_+_42697065 | 0.35 |
ENST00000565559.1 |
CAPN3 |
calpain 3, (p94) |
| chr6_+_158733692 | 0.35 |
ENST00000367094.2 ENST00000367097.3 |
TULP4 |
tubby like protein 4 |
| chr1_-_144994909 | 0.35 |
ENST00000369347.4 ENST00000369354.3 |
PDE4DIP |
phosphodiesterase 4D interacting protein |
| chr1_+_236305826 | 0.34 |
ENST00000366592.3 ENST00000366591.4 |
GPR137B |
G protein-coupled receptor 137B |
| chr1_+_15479021 | 0.34 |
ENST00000428417.1 |
TMEM51 |
transmembrane protein 51 |
| chr13_-_33112899 | 0.34 |
ENST00000267068.3 ENST00000357505.6 ENST00000399396.3 |
N4BP2L2 |
NEDD4 binding protein 2-like 2 |
| chr11_-_66445219 | 0.32 |
ENST00000525754.1 ENST00000531969.1 ENST00000524637.1 ENST00000531036.2 ENST00000310046.4 |
RBM4B |
RNA binding motif protein 4B |
| chr3_+_183894566 | 0.30 |
ENST00000439647.1 |
AP2M1 |
adaptor-related protein complex 2, mu 1 subunit |
| chr3_+_33840050 | 0.30 |
ENST00000457054.2 ENST00000413073.1 |
PDCD6IP |
programmed cell death 6 interacting protein |
| chr6_-_106773610 | 0.30 |
ENST00000369076.3 ENST00000369070.1 |
ATG5 |
autophagy related 5 |
| chr5_+_66124590 | 0.29 |
ENST00000490016.2 ENST00000403666.1 ENST00000450827.1 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
| chr8_-_59412717 | 0.29 |
ENST00000301645.3 |
CYP7A1 |
cytochrome P450, family 7, subfamily A, polypeptide 1 |
| chr17_-_7167279 | 0.28 |
ENST00000571932.2 |
CLDN7 |
claudin 7 |
| chr1_+_15479054 | 0.28 |
ENST00000376014.3 ENST00000451326.2 |
TMEM51 |
transmembrane protein 51 |
| chr17_-_27503770 | 0.28 |
ENST00000533112.1 |
MYO18A |
myosin XVIIIA |
| chr7_-_74221288 | 0.28 |
ENST00000451013.2 |
GTF2IRD2 |
GTF2I repeat domain containing 2 |
| chr16_-_47495170 | 0.28 |
ENST00000320640.6 ENST00000544001.2 |
ITFG1 |
integrin alpha FG-GAP repeat containing 1 |
| chr12_-_27167233 | 0.27 |
ENST00000535819.1 ENST00000543803.1 ENST00000535423.1 ENST00000539741.1 ENST00000343028.4 ENST00000545600.1 ENST00000543088.1 |
TM7SF3 |
transmembrane 7 superfamily member 3 |
| chr1_+_22962948 | 0.27 |
ENST00000374642.3 |
C1QA |
complement component 1, q subcomponent, A chain |
| chr18_+_3252206 | 0.27 |
ENST00000578562.2 |
MYL12A |
myosin, light chain 12A, regulatory, non-sarcomeric |
Gene Ontology Analysis
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 13.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 4.5 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.2 | 10.7 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.2 | 2.9 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 5.8 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 2.5 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 9.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 1.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 3.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 1.9 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.9 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 1.1 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 2.9 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 2.0 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 3.1 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.7 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.7 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.9 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 1.4 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 3.7 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.6 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 1.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.3 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.4 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 4.5 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.1 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 13.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.3 | 3.8 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.2 | 3.7 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.2 | 5.1 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 2.8 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 3.3 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 2.9 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 1.0 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 11.1 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 4.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 2.6 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.1 | 1.8 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 0.6 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 5.6 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 0.9 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 1.1 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 1.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.3 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.6 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.8 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.5 | REACTOME SIGNALING BY NOTCH2 | Genes involved in Signaling by NOTCH2 |
| 0.0 | 0.9 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.9 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.8 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.6 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.5 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 1.5 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.2 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.0 | 0.5 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 1.2 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.8 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.5 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 1.9 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.9 | 13.1 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.4 | 5.1 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.4 | 5.0 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.4 | 6.1 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.3 | 2.7 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.3 | 3.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) sorting endosome(GO:0097443) |
| 0.2 | 3.7 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 0.9 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.2 | 2.9 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.2 | 0.7 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.2 | 1.3 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.2 | 1.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.2 | 1.7 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 2.8 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 4.5 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 3.7 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 0.7 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.1 | 0.9 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 0.9 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 7.4 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.1 | 1.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 1.2 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 0.3 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.1 | 5.9 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 0.4 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.1 | 0.8 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 1.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 1.9 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 1.3 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.5 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.7 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 4.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.9 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.2 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.3 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.5 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.4 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.3 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 1.9 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 3.0 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 11.9 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 3.1 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 0.6 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 1.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 1.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 7.8 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 2.0 | GO:0098802 | plasma membrane receptor complex(GO:0098802) |
| 0.0 | 0.2 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.6 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 2.3 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.4 | GO:0048786 | presynaptic active zone(GO:0048786) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.6 | 2.5 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.6 | 5.0 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.6 | 1.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.5 | 1.5 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.4 | 2.4 | GO:0038049 | transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) |
| 0.4 | 3.2 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.3 | 2.5 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.3 | 2.8 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.2 | 1.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.2 | 2.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.2 | 6.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 0.6 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.2 | 17.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.2 | 0.7 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.2 | 4.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 1.0 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.2 | 2.9 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.2 | 0.8 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.2 | 5.8 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 1.3 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.6 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.1 | 1.2 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.1 | 1.2 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.1 | 3.7 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.9 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 0.7 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 0.7 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.1 | 0.9 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 0.3 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.1 | 1.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.1 | 2.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.9 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.1 | 1.2 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.6 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.1 | 0.5 | GO:0051022 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.1 | 16.7 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 0.3 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.1 | 0.5 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.3 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.1 | 0.4 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.1 | 0.7 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 0.8 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.1 | 2.1 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 1.3 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.9 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.5 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.2 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.5 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 1.5 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.4 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 3.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 7.4 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.8 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.2 | GO:0016301 | kinase activity(GO:0016301) |
| 0.0 | 1.3 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.7 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.3 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.2 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.3 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.8 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.5 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 1.9 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 1.0 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.7 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.4 | GO:0008200 | ion channel inhibitor activity(GO:0008200) |
| 0.0 | 0.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.0 | 2.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.0 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.0 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.0 | 0.7 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 3.6 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.4 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.5 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.8 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.4 | GO:0070888 | E-box binding(GO:0070888) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.0 | GO:0002605 | negative regulation of dendritic cell antigen processing and presentation(GO:0002605) |
| 1.5 | 4.5 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 1.2 | 4.8 | GO:0009956 | radial pattern formation(GO:0009956) |
| 1.1 | 13.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 1.0 | 5.0 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.9 | 3.6 | GO:0003274 | endocardial cushion fusion(GO:0003274) |
| 0.8 | 3.3 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.7 | 5.1 | GO:2000795 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.7 | 2.7 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.6 | 3.2 | GO:0021649 | vestibulocochlear nerve structural organization(GO:0021649) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) endothelial tip cell fate specification(GO:0097102) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.4 | 2.1 | GO:2000035 | positive regulation of somatic stem cell population maintenance(GO:1904674) regulation of stem cell division(GO:2000035) |
| 0.4 | 1.2 | GO:0014876 | response to injury involved in regulation of muscle adaptation(GO:0014876) |
| 0.4 | 6.6 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.4 | 2.9 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.3 | 1.7 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.3 | 1.0 | GO:0060940 | epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 0.3 | 1.3 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.3 | 0.9 | GO:0002384 | hepatic immune response(GO:0002384) response to prolactin(GO:1990637) regulation of STAT protein import into nucleus(GO:2000364) positive regulation of STAT protein import into nucleus(GO:2000366) |
| 0.3 | 1.7 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.3 | 5.1 | GO:0001765 | membrane raft assembly(GO:0001765) |
| 0.2 | 1.2 | GO:0051946 | regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 0.2 | 2.6 | GO:0061314 | Notch signaling involved in heart development(GO:0061314) |
| 0.2 | 0.7 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.2 | 1.3 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.2 | 1.3 | GO:0006789 | bilirubin conjugation(GO:0006789) |
| 0.2 | 4.3 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 0.6 | GO:0036511 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.2 | 5.2 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.2 | 0.6 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.2 | 0.6 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) |
| 0.2 | 1.3 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.2 | 0.7 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.2 | 0.5 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 0.2 | 10.3 | GO:0003254 | regulation of membrane depolarization(GO:0003254) |
| 0.2 | 0.5 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.2 | 0.5 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.2 | 0.9 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.2 | 0.8 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.2 | 0.3 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.1 | 0.4 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.1 | 0.6 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.1 | 2.8 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.1 | 1.1 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.1 | 1.7 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.1 | 0.3 | GO:0036378 | calcitriol biosynthetic process from calciol(GO:0036378) |
| 0.1 | 0.6 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.1 | 0.6 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.1 | 1.1 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.1 | 0.6 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.1 | 0.4 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.1 | 0.7 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.1 | 0.3 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.1 | 0.4 | GO:1905071 | ossification involved in bone remodeling(GO:0043932) frontal suture morphogenesis(GO:0060364) occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 0.1 | 0.9 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 3.9 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 0.9 | GO:1904261 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.1 | 0.5 | GO:0072719 | cellular response to cisplatin(GO:0072719) |
| 0.1 | 0.5 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.3 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
| 0.1 | 1.3 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.1 | 0.4 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.1 | 0.2 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.1 | 1.0 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.1 | 0.4 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 1.7 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.1 | 0.5 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 | 2.8 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 0.6 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.1 | 0.2 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) cellular alkene metabolic process(GO:0043449) |
| 0.1 | 1.0 | GO:0043687 | post-translational protein modification(GO:0043687) |
| 0.1 | 0.7 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.1 | 2.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 0.9 | GO:0051683 | establishment of Golgi localization(GO:0051683) |
| 0.0 | 0.8 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.7 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.3 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.0 | 0.5 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.5 | GO:0035376 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.0 | 0.4 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 1.0 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 1.0 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.3 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.7 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 | 0.9 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.5 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.4 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 0.0 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.2 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.0 | 0.0 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.0 | 1.7 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 0.9 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.0 | 0.5 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.5 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.5 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 1.2 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.1 | GO:0002933 | lipid hydroxylation(GO:0002933) omega-hydroxylase P450 pathway(GO:0097267) |
| 0.0 | 0.2 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.0 | 0.4 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 1.1 | GO:0000305 | response to oxygen radical(GO:0000305) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 1.9 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 | 0.4 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.3 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 0.0 | 0.1 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.2 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 2.0 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 0.9 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.5 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.0 | 0.0 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.5 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 | 0.1 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.7 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.0 | 0.2 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.2 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.3 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.0 | 0.1 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.0 | 0.3 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.3 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.0 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 | 0.3 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 3.8 | GO:0007517 | muscle organ development(GO:0007517) |
| 0.0 | 0.3 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.5 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 1.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.4 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.0 | 0.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |