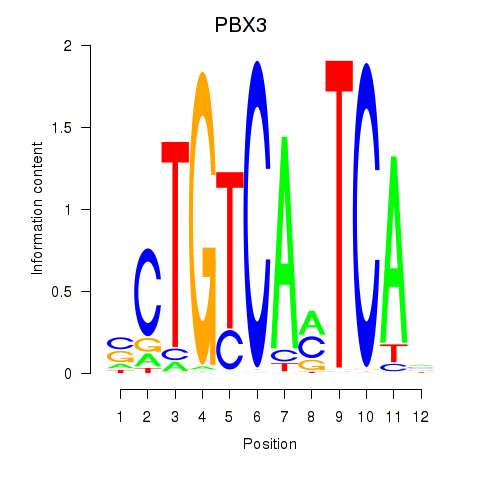
Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for PBX3
Z-value: 1.40
Transcription factors associated with PBX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PBX3
|
ENSG00000167081.12 | PBX3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PBX3 | hg19_v2_chr9_+_128509663_128509733 | 0.10 | 7.0e-01 | Click! |
Activity profile of PBX3 motif
Sorted Z-values of PBX3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of PBX3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_89238149 | 6.58 |
ENST00000289746.2 |
CDH15 |
cadherin 15, type 1, M-cadherin (myotubule) |
| chr2_-_179672142 | 4.41 |
ENST00000342992.6 ENST00000360870.5 ENST00000460472.2 ENST00000589042.1 ENST00000591111.1 ENST00000342175.6 ENST00000359218.5 |
TTN |
titin |
| chr11_-_2170786 | 2.78 |
ENST00000300632.5 |
IGF2 |
insulin-like growth factor 2 (somatomedin A) |
| chr2_-_45236540 | 2.34 |
ENST00000303077.6 |
SIX2 |
SIX homeobox 2 |
| chr9_-_35689900 | 2.13 |
ENST00000378300.5 ENST00000329305.2 ENST00000360958.2 |
TPM2 |
tropomyosin 2 (beta) |
| chr17_-_1619535 | 2.08 |
ENST00000573075.1 ENST00000574306.1 |
MIR22HG |
MIR22 host gene (non-protein coding) |
| chrX_+_100805496 | 2.06 |
ENST00000372829.3 |
ARMCX1 |
armadillo repeat containing, X-linked 1 |
| chr3_+_8775466 | 2.04 |
ENST00000343849.2 ENST00000397368.2 |
CAV3 |
caveolin 3 |
| chr17_-_1619491 | 1.99 |
ENST00000570416.1 ENST00000575626.1 ENST00000610106.1 ENST00000608198.1 ENST00000609442.1 ENST00000334146.3 ENST00000576489.1 ENST00000608245.1 ENST00000609398.1 ENST00000608913.1 ENST00000574016.1 ENST00000571091.1 ENST00000573127.1 ENST00000609990.1 ENST00000576749.1 |
MIR22HG |
MIR22 host gene (non-protein coding) |
| chrX_-_100914781 | 1.90 |
ENST00000431597.1 ENST00000458024.1 ENST00000413506.1 ENST00000440675.1 ENST00000328766.5 ENST00000356824.4 |
ARMCX2 |
armadillo repeat containing, X-linked 2 |
| chr17_+_48243352 | 1.82 |
ENST00000344627.6 ENST00000262018.3 ENST00000543315.1 ENST00000451235.2 ENST00000511303.1 |
SGCA |
sarcoglycan, alpha (50kDa dystrophin-associated glycoprotein) |
| chr3_+_30647994 | 1.81 |
ENST00000295754.5 |
TGFBR2 |
transforming growth factor, beta receptor II (70/80kDa) |
| chr1_-_173176452 | 1.77 |
ENST00000281834.3 |
TNFSF4 |
tumor necrosis factor (ligand) superfamily, member 4 |
| chr3_+_30648066 | 1.73 |
ENST00000359013.4 |
TGFBR2 |
transforming growth factor, beta receptor II (70/80kDa) |
| chr3_-_52486841 | 1.73 |
ENST00000496590.1 |
TNNC1 |
troponin C type 1 (slow) |
| chr16_-_53537105 | 1.66 |
ENST00000568596.1 ENST00000570004.1 ENST00000564497.1 ENST00000300245.4 ENST00000394657.7 |
AKTIP |
AKT interacting protein |
| chr6_-_169654139 | 1.58 |
ENST00000366787.3 |
THBS2 |
thrombospondin 2 |
| chr9_+_75766652 | 1.56 |
ENST00000257497.6 |
ANXA1 |
annexin A1 |
| chr1_+_100111479 | 1.42 |
ENST00000263174.4 |
PALMD |
palmdelphin |
| chr10_-_76868931 | 1.42 |
ENST00000372700.3 ENST00000473072.2 ENST00000491677.2 ENST00000607131.1 ENST00000372702.3 |
DUSP13 |
dual specificity phosphatase 13 |
| chr1_-_110283660 | 1.42 |
ENST00000361066.2 |
GSTM3 |
glutathione S-transferase mu 3 (brain) |
| chr1_-_110283138 | 1.40 |
ENST00000256594.3 |
GSTM3 |
glutathione S-transferase mu 3 (brain) |
| chr2_+_20646824 | 1.36 |
ENST00000272233.4 |
RHOB |
ras homolog family member B |
| chrX_-_153637612 | 1.35 |
ENST00000369807.1 ENST00000369808.3 |
DNASE1L1 |
deoxyribonuclease I-like 1 |
| chr21_-_39288743 | 1.21 |
ENST00000609713.1 |
KCNJ6 |
potassium inwardly-rectifying channel, subfamily J, member 6 |
| chrX_+_102631248 | 1.20 |
ENST00000361298.4 ENST00000372645.3 ENST00000372635.1 |
NGFRAP1 |
nerve growth factor receptor (TNFRSF16) associated protein 1 |
| chr10_+_102790980 | 1.18 |
ENST00000393459.1 ENST00000224807.5 |
SFXN3 |
sideroflexin 3 |
| chr12_+_75874984 | 1.18 |
ENST00000550491.1 |
GLIPR1 |
GLI pathogenesis-related 1 |
| chr13_+_73632897 | 1.17 |
ENST00000377687.4 |
KLF5 |
Kruppel-like factor 5 (intestinal) |
| chr12_+_75874460 | 1.16 |
ENST00000266659.3 |
GLIPR1 |
GLI pathogenesis-related 1 |
| chr1_+_100111580 | 1.16 |
ENST00000605497.1 |
PALMD |
palmdelphin |
| chr11_+_112832202 | 1.13 |
ENST00000534015.1 |
NCAM1 |
neural cell adhesion molecule 1 |
| chr2_+_201171242 | 1.12 |
ENST00000360760.5 |
SPATS2L |
spermatogenesis associated, serine-rich 2-like |
| chr15_+_25068773 | 1.07 |
ENST00000400100.1 ENST00000400098.1 |
SNRPN |
small nuclear ribonucleoprotein polypeptide N |
| chr16_+_56642041 | 1.07 |
ENST00000245185.5 |
MT2A |
metallothionein 2A |
| chr5_-_179780312 | 1.06 |
ENST00000253778.8 |
GFPT2 |
glutamine-fructose-6-phosphate transaminase 2 |
| chr3_+_35681081 | 1.03 |
ENST00000428373.1 |
ARPP21 |
cAMP-regulated phosphoprotein, 21kDa |
| chr2_+_201171064 | 0.99 |
ENST00000451764.2 |
SPATS2L |
spermatogenesis associated, serine-rich 2-like |
| chr2_+_201171372 | 0.99 |
ENST00000409140.3 |
SPATS2L |
spermatogenesis associated, serine-rich 2-like |
| chr3_+_197518100 | 0.98 |
ENST00000438796.2 ENST00000414675.2 ENST00000441090.2 ENST00000334859.4 ENST00000425562.2 |
LRCH3 |
leucine-rich repeats and calponin homology (CH) domain containing 3 |
| chr19_+_16435625 | 0.96 |
ENST00000248071.5 ENST00000592003.1 |
KLF2 |
Kruppel-like factor 2 |
| chr12_+_75874580 | 0.94 |
ENST00000456650.3 |
GLIPR1 |
GLI pathogenesis-related 1 |
| chr19_-_45996465 | 0.94 |
ENST00000430715.2 |
RTN2 |
reticulon 2 |
| chr2_+_201170770 | 0.92 |
ENST00000409988.3 ENST00000409385.1 |
SPATS2L |
spermatogenesis associated, serine-rich 2-like |
| chr9_+_34652164 | 0.90 |
ENST00000441545.2 ENST00000553620.1 |
IL11RA |
interleukin 11 receptor, alpha |
| chr11_+_112832133 | 0.89 |
ENST00000524665.1 |
NCAM1 |
neural cell adhesion molecule 1 |
| chr9_+_82186872 | 0.84 |
ENST00000376544.3 ENST00000376520.4 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr9_+_116327326 | 0.84 |
ENST00000342620.5 |
RGS3 |
regulator of G-protein signaling 3 |
| chr12_-_91573249 | 0.82 |
ENST00000550099.1 ENST00000546391.1 ENST00000551354.1 |
DCN |
decorin |
| chr12_-_91573132 | 0.75 |
ENST00000550563.1 ENST00000546370.1 |
DCN |
decorin |
| chr10_+_114709999 | 0.73 |
ENST00000355995.4 ENST00000545257.1 ENST00000543371.1 ENST00000536810.1 ENST00000355717.4 ENST00000538897.1 ENST00000534894.1 |
TCF7L2 |
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr19_-_23578220 | 0.73 |
ENST00000595533.1 ENST00000397082.2 ENST00000599743.1 ENST00000300619.7 |
ZNF91 |
zinc finger protein 91 |
| chr5_-_140700322 | 0.72 |
ENST00000313368.5 |
TAF7 |
TAF7 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 55kDa |
| chr13_-_33780133 | 0.72 |
ENST00000399365.3 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
| chr7_+_106809406 | 0.71 |
ENST00000468410.1 ENST00000478930.1 ENST00000464009.1 ENST00000222574.4 |
HBP1 |
HMG-box transcription factor 1 |
| chr6_-_100912785 | 0.68 |
ENST00000369208.3 |
SIM1 |
single-minded family bHLH transcription factor 1 |
| chr11_+_842928 | 0.68 |
ENST00000397408.1 |
TSPAN4 |
tetraspanin 4 |
| chr19_-_49496557 | 0.68 |
ENST00000323798.3 ENST00000541188.1 ENST00000544287.1 ENST00000540532.1 ENST00000263276.6 |
GYS1 |
glycogen synthase 1 (muscle) |
| chr11_+_112832090 | 0.68 |
ENST00000533760.1 |
NCAM1 |
neural cell adhesion molecule 1 |
| chr12_-_91573316 | 0.67 |
ENST00000393155.1 |
DCN |
decorin |
| chr16_+_30210552 | 0.67 |
ENST00000338971.5 |
SULT1A3 |
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 |
| chr3_+_52489503 | 0.66 |
ENST00000345716.4 |
NISCH |
nischarin |
| chr17_+_39975455 | 0.63 |
ENST00000455106.1 |
FKBP10 |
FK506 binding protein 10, 65 kDa |
| chr10_+_28966271 | 0.60 |
ENST00000375533.3 |
BAMBI |
BMP and activin membrane-bound inhibitor |
| chr12_-_56123444 | 0.60 |
ENST00000546457.1 ENST00000549117.1 |
CD63 |
CD63 molecule |
| chr17_+_39975544 | 0.59 |
ENST00000544340.1 |
FKBP10 |
FK506 binding protein 10, 65 kDa |
| chr2_-_220435963 | 0.59 |
ENST00000373876.1 ENST00000404537.1 ENST00000603926.1 ENST00000373873.4 ENST00000289656.3 |
OBSL1 |
obscurin-like 1 |
| chr19_-_12405689 | 0.58 |
ENST00000355684.5 |
ZNF44 |
zinc finger protein 44 |
| chr7_-_30066233 | 0.58 |
ENST00000222803.5 |
FKBP14 |
FK506 binding protein 14, 22 kDa |
| chr19_-_14228541 | 0.57 |
ENST00000590853.1 ENST00000308677.4 |
PRKACA |
protein kinase, cAMP-dependent, catalytic, alpha |
| chr9_+_82186682 | 0.57 |
ENST00000376552.2 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr9_-_35115836 | 0.56 |
ENST00000378566.1 ENST00000378554.2 ENST00000322813.5 |
FAM214B |
family with sequence similarity 214, member B |
| chrX_-_134049262 | 0.55 |
ENST00000370783.3 |
MOSPD1 |
motile sperm domain containing 1 |
| chr6_+_41021027 | 0.54 |
ENST00000244669.2 |
APOBEC2 |
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2 |
| chr1_+_7831323 | 0.53 |
ENST00000054666.6 |
VAMP3 |
vesicle-associated membrane protein 3 |
| chr14_-_82000140 | 0.52 |
ENST00000555824.1 ENST00000557372.1 ENST00000336735.4 |
SEL1L |
sel-1 suppressor of lin-12-like (C. elegans) |
| chr7_+_150929550 | 0.50 |
ENST00000482173.1 ENST00000495645.1 ENST00000035307.2 |
CHPF2 |
chondroitin polymerizing factor 2 |
| chr11_+_842808 | 0.50 |
ENST00000397397.2 ENST00000397411.2 ENST00000397396.1 |
TSPAN4 |
tetraspanin 4 |
| chr20_+_35807512 | 0.48 |
ENST00000373622.5 |
RPN2 |
ribophorin II |
| chr19_+_21106081 | 0.47 |
ENST00000300540.3 ENST00000595854.1 ENST00000601284.1 ENST00000328178.8 ENST00000599885.1 ENST00000596476.1 ENST00000345030.6 |
ZNF85 |
zinc finger protein 85 |
| chr10_-_101945771 | 0.47 |
ENST00000370408.2 ENST00000407654.3 |
ERLIN1 |
ER lipid raft associated 1 |
| chr3_+_101498269 | 0.46 |
ENST00000491511.2 |
NXPE3 |
neurexophilin and PC-esterase domain family, member 3 |
| chr5_-_150460914 | 0.46 |
ENST00000389378.2 |
TNIP1 |
TNFAIP3 interacting protein 1 |
| chrX_+_102883887 | 0.45 |
ENST00000372625.3 ENST00000372624.3 |
TCEAL1 |
transcription elongation factor A (SII)-like 1 |
| chr4_+_6271558 | 0.45 |
ENST00000503569.1 ENST00000226760.1 |
WFS1 |
Wolfram syndrome 1 (wolframin) |
| chr17_+_46125707 | 0.44 |
ENST00000584137.1 ENST00000362042.3 ENST00000585291.1 ENST00000357480.5 |
NFE2L1 |
nuclear factor, erythroid 2-like 1 |
| chr22_-_37172111 | 0.44 |
ENST00000417951.2 ENST00000430701.1 ENST00000433985.2 |
IFT27 |
intraflagellar transport 27 homolog (Chlamydomonas) |
| chr10_+_114710211 | 0.44 |
ENST00000349937.2 ENST00000369397.4 |
TCF7L2 |
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr20_+_35807449 | 0.43 |
ENST00000237530.6 |
RPN2 |
ribophorin II |
| chr19_+_40877583 | 0.43 |
ENST00000596470.1 |
PLD3 |
phospholipase D family, member 3 |
| chr22_-_37172191 | 0.41 |
ENST00000340630.5 |
IFT27 |
intraflagellar transport 27 homolog (Chlamydomonas) |
| chr4_+_90800656 | 0.41 |
ENST00000394980.1 |
MMRN1 |
multimerin 1 |
| chr7_-_38370536 | 0.40 |
ENST00000390343.2 |
TRGV8 |
T cell receptor gamma variable 8 |
| chr21_-_30257669 | 0.40 |
ENST00000303775.5 ENST00000351429.3 |
N6AMT1 |
N-6 adenine-specific DNA methyltransferase 1 (putative) |
| chr3_-_88108212 | 0.39 |
ENST00000482016.1 |
CGGBP1 |
CGG triplet repeat binding protein 1 |
| chr19_-_50063907 | 0.38 |
ENST00000598296.1 |
NOSIP |
nitric oxide synthase interacting protein |
| chr16_+_67927147 | 0.38 |
ENST00000291041.5 |
PSKH1 |
protein serine kinase H1 |
| chr19_+_44556158 | 0.38 |
ENST00000434772.3 ENST00000585552.1 |
ZNF223 |
zinc finger protein 223 |
| chr6_-_131321863 | 0.37 |
ENST00000528282.1 |
EPB41L2 |
erythrocyte membrane protein band 4.1-like 2 |
| chr16_-_30032610 | 0.37 |
ENST00000574405.1 |
DOC2A |
double C2-like domains, alpha |
| chr10_+_99258625 | 0.37 |
ENST00000370664.3 |
UBTD1 |
ubiquitin domain containing 1 |
| chr4_+_8594364 | 0.36 |
ENST00000360986.4 |
CPZ |
carboxypeptidase Z |
| chr6_+_110299501 | 0.36 |
ENST00000414000.2 |
GPR6 |
G protein-coupled receptor 6 |
| chr19_-_4867665 | 0.35 |
ENST00000592528.1 ENST00000589494.1 ENST00000585479.1 ENST00000221957.4 |
PLIN3 |
perilipin 3 |
| chr6_-_32145861 | 0.35 |
ENST00000336984.6 |
AGPAT1 |
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr3_+_15643140 | 0.35 |
ENST00000449107.1 |
BTD |
biotinidase |
| chr19_+_4969116 | 0.34 |
ENST00000588337.1 ENST00000159111.4 ENST00000381759.4 |
KDM4B |
lysine (K)-specific demethylase 4B |
| chr7_+_116502605 | 0.34 |
ENST00000458284.2 ENST00000490693.1 |
CAPZA2 |
capping protein (actin filament) muscle Z-line, alpha 2 |
| chr17_-_13505219 | 0.34 |
ENST00000284110.1 |
HS3ST3A1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 |
| chr7_+_99699179 | 0.33 |
ENST00000438383.1 ENST00000429084.1 ENST00000359593.4 ENST00000439416.1 |
AP4M1 |
adaptor-related protein complex 4, mu 1 subunit |
| chr3_+_184279566 | 0.33 |
ENST00000330394.2 |
EPHB3 |
EPH receptor B3 |
| chr22_-_39151947 | 0.33 |
ENST00000216064.4 |
SUN2 |
Sad1 and UNC84 domain containing 2 |
| chr19_+_38924316 | 0.33 |
ENST00000355481.4 ENST00000360985.3 ENST00000359596.3 |
RYR1 |
ryanodine receptor 1 (skeletal) |
| chr1_-_155211017 | 0.32 |
ENST00000536770.1 ENST00000368373.3 |
GBA |
glucosidase, beta, acid |
| chr7_+_99699280 | 0.32 |
ENST00000421755.1 |
AP4M1 |
adaptor-related protein complex 4, mu 1 subunit |
| chr19_-_12251202 | 0.31 |
ENST00000334213.5 |
ZNF20 |
zinc finger protein 20 |
| chr17_+_58755184 | 0.31 |
ENST00000589222.1 ENST00000407086.3 ENST00000390652.5 |
BCAS3 |
breast carcinoma amplified sequence 3 |
| chr14_-_24584138 | 0.31 |
ENST00000558280.1 ENST00000561028.1 |
NRL |
neural retina leucine zipper |
| chr14_-_91526922 | 0.30 |
ENST00000418736.2 ENST00000261991.3 |
RPS6KA5 |
ribosomal protein S6 kinase, 90kDa, polypeptide 5 |
| chr1_+_6845384 | 0.30 |
ENST00000303635.7 |
CAMTA1 |
calmodulin binding transcription activator 1 |
| chr7_+_116502527 | 0.30 |
ENST00000361183.3 |
CAPZA2 |
capping protein (actin filament) muscle Z-line, alpha 2 |
| chr5_+_149569520 | 0.30 |
ENST00000230671.2 ENST00000524041.1 |
SLC6A7 |
solute carrier family 6 (neurotransmitter transporter), member 7 |
| chr3_+_15643245 | 0.30 |
ENST00000303498.5 ENST00000437172.1 |
BTD |
biotinidase |
| chr12_+_51632508 | 0.30 |
ENST00000449723.3 |
DAZAP2 |
DAZ associated protein 2 |
| chr3_-_88108192 | 0.29 |
ENST00000309534.6 |
CGGBP1 |
CGG triplet repeat binding protein 1 |
| chr7_+_134331550 | 0.29 |
ENST00000344924.3 ENST00000418040.1 ENST00000393132.2 |
BPGM |
2,3-bisphosphoglycerate mutase |
| chr22_-_39151995 | 0.29 |
ENST00000405018.1 ENST00000438058.1 |
SUN2 |
Sad1 and UNC84 domain containing 2 |
| chr3_+_15643476 | 0.28 |
ENST00000436193.1 ENST00000383778.4 |
BTD |
biotinidase |
| chr10_-_79789291 | 0.28 |
ENST00000372371.3 |
POLR3A |
polymerase (RNA) III (DNA directed) polypeptide A, 155kDa |
| chr4_+_124317940 | 0.27 |
ENST00000505319.1 ENST00000339241.1 |
SPRY1 |
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
| chr9_+_34989638 | 0.27 |
ENST00000453597.3 ENST00000335998.3 ENST00000312316.5 |
DNAJB5 |
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chr9_-_128003606 | 0.27 |
ENST00000324460.6 |
HSPA5 |
heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa) |
| chr8_-_41522779 | 0.27 |
ENST00000522231.1 ENST00000314214.8 ENST00000348036.4 ENST00000457297.1 ENST00000522543.1 |
ANK1 |
ankyrin 1, erythrocytic |
| chr1_-_155211065 | 0.27 |
ENST00000427500.3 |
GBA |
glucosidase, beta, acid |
| chr16_+_31044413 | 0.27 |
ENST00000394998.1 |
STX4 |
syntaxin 4 |
| chr5_-_37249397 | 0.27 |
ENST00000425232.2 ENST00000274258.7 |
C5orf42 |
chromosome 5 open reading frame 42 |
| chr7_-_100183742 | 0.25 |
ENST00000310300.6 |
LRCH4 |
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr20_-_34330129 | 0.25 |
ENST00000397370.3 ENST00000528062.3 ENST00000407261.4 ENST00000374038.3 ENST00000361162.6 |
RBM39 |
RNA binding motif protein 39 |
| chr6_-_11382478 | 0.25 |
ENST00000397378.3 ENST00000513989.1 ENST00000508546.1 ENST00000504387.1 |
NEDD9 |
neural precursor cell expressed, developmentally down-regulated 9 |
| chr10_-_44144152 | 0.24 |
ENST00000395797.1 |
ZNF32 |
zinc finger protein 32 |
| chr19_+_2841433 | 0.24 |
ENST00000334241.4 ENST00000585966.1 ENST00000591539.1 |
ZNF555 |
zinc finger protein 555 |
| chr19_+_12035913 | 0.24 |
ENST00000591944.1 |
ZNF763 |
Uncharacterized protein; Zinc finger protein 763 |
| chr16_+_85061367 | 0.24 |
ENST00000538274.1 ENST00000258180.3 |
KIAA0513 |
KIAA0513 |
| chr3_-_49823941 | 0.24 |
ENST00000321599.4 ENST00000395238.1 ENST00000468463.1 ENST00000460540.1 |
IP6K1 |
inositol hexakisphosphate kinase 1 |
| chr17_+_61699766 | 0.24 |
ENST00000579585.1 ENST00000584573.1 ENST00000361733.3 ENST00000361357.3 |
MAP3K3 |
mitogen-activated protein kinase kinase kinase 3 |
| chr12_-_16761007 | 0.24 |
ENST00000354662.1 ENST00000441439.2 |
LMO3 |
LIM domain only 3 (rhombotin-like 2) |
| chr19_-_45873642 | 0.23 |
ENST00000485403.2 ENST00000586856.1 ENST00000586131.1 ENST00000391940.4 ENST00000221481.6 ENST00000391944.3 ENST00000391945.4 |
ERCC2 |
excision repair cross-complementing rodent repair deficiency, complementation group 2 |
| chr19_+_55591743 | 0.23 |
ENST00000588359.1 ENST00000245618.5 |
EPS8L1 |
EPS8-like 1 |
| chr11_-_86383157 | 0.23 |
ENST00000393324.3 |
ME3 |
malic enzyme 3, NADP(+)-dependent, mitochondrial |
| chr19_+_21688366 | 0.23 |
ENST00000358491.4 ENST00000597078.1 |
ZNF429 |
zinc finger protein 429 |
| chr22_-_36784035 | 0.23 |
ENST00000216181.5 |
MYH9 |
myosin, heavy chain 9, non-muscle |
| chr2_-_71062938 | 0.22 |
ENST00000410009.3 |
CD207 |
CD207 molecule, langerin |
| chr17_-_17875688 | 0.22 |
ENST00000379504.3 ENST00000318094.10 ENST00000540946.1 ENST00000542206.1 ENST00000395739.4 ENST00000581396.1 ENST00000535933.1 ENST00000579586.1 |
TOM1L2 |
target of myb1-like 2 (chicken) |
| chr8_+_26240414 | 0.22 |
ENST00000380629.2 |
BNIP3L |
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr17_+_17876127 | 0.22 |
ENST00000582416.1 ENST00000313838.8 ENST00000411504.2 ENST00000581264.1 ENST00000399187.1 ENST00000479684.2 ENST00000584166.1 ENST00000585108.1 ENST00000399182.1 ENST00000579977.1 |
LRRC48 |
leucine rich repeat containing 48 |
| chr2_-_220094031 | 0.22 |
ENST00000443140.1 ENST00000432520.1 ENST00000409618.1 |
ATG9A |
autophagy related 9A |
| chr1_-_28969517 | 0.22 |
ENST00000263974.4 ENST00000373824.4 |
TAF12 |
TAF12 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 20kDa |
| chr3_+_48481658 | 0.21 |
ENST00000438607.2 |
TMA7 |
translation machinery associated 7 homolog (S. cerevisiae) |
| chr17_-_8066250 | 0.21 |
ENST00000488857.1 ENST00000481878.1 ENST00000316509.6 ENST00000498285.1 |
VAMP2 RP11-599B13.6 |
vesicle-associated membrane protein 2 (synaptobrevin 2) Uncharacterized protein |
| chr14_+_23938891 | 0.21 |
ENST00000408901.3 ENST00000397154.3 ENST00000555128.1 |
NGDN |
neuroguidin, EIF4E binding protein |
| chr11_-_86383370 | 0.21 |
ENST00000526834.1 ENST00000359636.2 |
ME3 |
malic enzyme 3, NADP(+)-dependent, mitochondrial |
| chr10_-_44144292 | 0.21 |
ENST00000374433.2 |
ZNF32 |
zinc finger protein 32 |
| chr16_-_79634595 | 0.21 |
ENST00000326043.4 ENST00000393350.1 |
MAF |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog |
| chr7_+_72742162 | 0.21 |
ENST00000431982.2 |
FKBP6 |
FK506 binding protein 6, 36kDa |
| chr14_+_24583836 | 0.21 |
ENST00000559115.1 ENST00000558215.1 ENST00000557810.1 ENST00000561375.1 ENST00000446197.3 ENST00000559796.1 ENST00000560713.1 ENST00000560901.1 ENST00000559382.1 |
DCAF11 |
DDB1 and CUL4 associated factor 11 |
| chr21_+_44394620 | 0.21 |
ENST00000291547.5 |
PKNOX1 |
PBX/knotted 1 homeobox 1 |
| chr13_+_20532848 | 0.20 |
ENST00000382874.2 |
ZMYM2 |
zinc finger, MYM-type 2 |
| chr12_-_56615693 | 0.20 |
ENST00000394013.2 ENST00000345093.4 ENST00000551711.1 ENST00000552656.1 |
RNF41 |
ring finger protein 41 |
| chr12_+_51632638 | 0.20 |
ENST00000549732.2 |
DAZAP2 |
DAZ associated protein 2 |
| chr7_+_72742178 | 0.20 |
ENST00000442793.1 ENST00000413573.2 ENST00000252037.4 |
FKBP6 |
FK506 binding protein 6, 36kDa |
| chr16_+_1756162 | 0.20 |
ENST00000250894.4 ENST00000356010.5 |
MAPK8IP3 |
mitogen-activated protein kinase 8 interacting protein 3 |
| chr7_+_64838712 | 0.19 |
ENST00000328747.7 ENST00000431504.1 ENST00000357512.2 |
ZNF92 |
zinc finger protein 92 |
| chr19_-_23941639 | 0.19 |
ENST00000395385.3 ENST00000531570.1 ENST00000528059.1 |
ZNF681 |
zinc finger protein 681 |
| chr20_-_17539456 | 0.19 |
ENST00000544874.1 ENST00000377868.2 |
BFSP1 |
beaded filament structural protein 1, filensin |
| chr13_+_20532900 | 0.19 |
ENST00000382871.2 |
ZMYM2 |
zinc finger, MYM-type 2 |
| chr7_+_149571045 | 0.18 |
ENST00000479613.1 ENST00000606024.1 ENST00000464662.1 ENST00000425642.2 |
ATP6V0E2 |
ATPase, H+ transporting V0 subunit e2 |
| chr19_+_41284121 | 0.18 |
ENST00000594800.1 ENST00000357052.2 ENST00000602173.1 |
RAB4B |
RAB4B, member RAS oncogene family |
| chr2_-_220436248 | 0.18 |
ENST00000265318.4 |
OBSL1 |
obscurin-like 1 |
| chr14_+_76127529 | 0.18 |
ENST00000556977.1 ENST00000557636.1 ENST00000286650.5 ENST00000298832.9 |
TTLL5 |
tubulin tyrosine ligase-like family, member 5 |
| chr11_-_73720122 | 0.18 |
ENST00000426995.2 |
UCP3 |
uncoupling protein 3 (mitochondrial, proton carrier) |
| chrX_+_153059608 | 0.17 |
ENST00000370087.1 |
SSR4 |
signal sequence receptor, delta |
| chr14_-_73925225 | 0.17 |
ENST00000356296.4 ENST00000355058.3 ENST00000359560.3 ENST00000557597.1 ENST00000554394.1 ENST00000555238.1 ENST00000535282.1 ENST00000555987.1 ENST00000555394.1 ENST00000554546.1 |
NUMB |
numb homolog (Drosophila) |
| chr17_-_40306934 | 0.17 |
ENST00000592574.1 ENST00000550406.1 ENST00000547517.1 ENST00000393860.3 ENST00000346213.4 |
CTD-2132N18.3 RAB5C |
Uncharacterized protein RAB5C, member RAS oncogene family |
| chr12_+_123717458 | 0.17 |
ENST00000253233.1 |
C12orf65 |
chromosome 12 open reading frame 65 |
| chr8_-_41522719 | 0.17 |
ENST00000335651.6 |
ANK1 |
ankyrin 1, erythrocytic |
| chr19_-_57988871 | 0.17 |
ENST00000596831.1 ENST00000601768.1 ENST00000356584.3 ENST00000600175.1 ENST00000425074.3 ENST00000343280.4 ENST00000427512.2 |
AC004076.9 ZNF772 |
Uncharacterized protein zinc finger protein 772 |
| chr8_+_31496809 | 0.16 |
ENST00000518104.1 ENST00000519301.1 |
NRG1 |
neuregulin 1 |
| chr7_+_55433131 | 0.16 |
ENST00000254770.2 |
LANCL2 |
LanC lantibiotic synthetase component C-like 2 (bacterial) |
| chr17_-_8534031 | 0.16 |
ENST00000411957.1 ENST00000396239.1 ENST00000379980.4 |
MYH10 |
myosin, heavy chain 10, non-muscle |
| chr7_-_38407770 | 0.16 |
ENST00000390348.2 |
TRGV1 |
T cell receptor gamma variable 1 (non-functional) |
| chr7_-_75368248 | 0.16 |
ENST00000434438.2 ENST00000336926.6 |
HIP1 |
huntingtin interacting protein 1 |
| chr16_-_89556942 | 0.16 |
ENST00000301030.4 |
ANKRD11 |
ankyrin repeat domain 11 |
| chr3_-_187454281 | 0.16 |
ENST00000232014.4 |
BCL6 |
B-cell CLL/lymphoma 6 |
| chr1_+_205197304 | 0.16 |
ENST00000358024.3 |
TMCC2 |
transmembrane and coiled-coil domain family 2 |
| chr22_-_39151463 | 0.16 |
ENST00000405510.1 ENST00000433561.1 |
SUN2 |
Sad1 and UNC84 domain containing 2 |
| chr16_+_28889703 | 0.15 |
ENST00000357084.3 |
ATP2A1 |
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr9_-_33264557 | 0.15 |
ENST00000473781.1 ENST00000488499.1 |
BAG1 |
BCL2-associated athanogene |
| chr2_-_220094294 | 0.15 |
ENST00000436856.1 ENST00000428226.1 ENST00000409422.1 ENST00000431715.1 ENST00000457841.1 ENST00000439812.1 ENST00000361242.4 ENST00000396761.2 |
ATG9A |
autophagy related 9A |
| chr17_+_7482785 | 0.15 |
ENST00000250092.6 ENST00000380498.6 ENST00000584502.1 |
CD68 |
CD68 molecule |
| chr19_+_21265028 | 0.15 |
ENST00000291770.7 |
ZNF714 |
zinc finger protein 714 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.4 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 1.2 | 3.5 | GO:0002663 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.8 | 2.3 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.6 | 1.8 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.6 | 2.8 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.5 | 1.6 | GO:1900138 | negative regulation of phospholipase A2 activity(GO:1900138) |
| 0.4 | 1.7 | GO:0032972 | diaphragm contraction(GO:0002086) regulation of muscle filament sliding speed(GO:0032972) |
| 0.4 | 1.2 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.3 | 1.0 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.3 | 2.8 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.2 | 1.4 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.2 | 1.1 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.2 | 2.2 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.2 | 1.8 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.2 | 0.5 | GO:0052250 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.1 | 0.6 | GO:1901804 | beta-glucoside metabolic process(GO:1901804) beta-glucoside catabolic process(GO:1901805) positive regulation of neuronal action potential(GO:1904457) |
| 0.1 | 0.7 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 0.3 | GO:0097501 | stress response to metal ion(GO:0097501) |
| 0.1 | 1.2 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.1 | 0.8 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.1 | 0.4 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.1 | 0.7 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 0.1 | 0.3 | GO:0031448 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.1 | 0.7 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.1 | 0.5 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 | 0.3 | GO:0060940 | epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 0.1 | 0.5 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.3 | GO:0043311 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.1 | 0.2 | GO:0021592 | fourth ventricle development(GO:0021592) third ventricle development(GO:0021678) |
| 0.1 | 0.2 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.1 | 0.3 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.1 | 1.1 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.1 | 1.4 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.1 | 0.2 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) |
| 0.1 | 0.7 | GO:0015846 | polyamine transport(GO:0015846) |
| 0.1 | 0.3 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.1 | 0.8 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.6 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.1 | 0.3 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.1 | 0.3 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) histone H2A phosphorylation(GO:1990164) |
| 0.1 | 0.2 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) |
| 0.1 | 0.7 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 0.2 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.1 | 6.5 | GO:0051149 | positive regulation of muscle cell differentiation(GO:0051149) |
| 0.1 | 0.4 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.4 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.0 | 0.3 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.4 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.0 | 0.9 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.0 | 0.6 | GO:0035826 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.0 | 0.1 | GO:0071545 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) inositol phosphate catabolic process(GO:0071545) |
| 0.0 | 1.7 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.0 | 0.2 | GO:0010641 | positive regulation of platelet-derived growth factor receptor signaling pathway(GO:0010641) |
| 0.0 | 0.2 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.2 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.7 | GO:0048243 | norepinephrine secretion(GO:0048243) |
| 0.0 | 0.7 | GO:0045199 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.5 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.0 | 0.6 | GO:0035413 | positive regulation of catenin import into nucleus(GO:0035413) |
| 0.0 | 0.2 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.1 | GO:0044857 | plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.3 | GO:0043931 | ossification involved in bone maturation(GO:0043931) |
| 0.0 | 2.1 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 1.4 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 0.6 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.3 | GO:0030262 | apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.4 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.1 | GO:1902617 | response to fluoride(GO:1902617) |
| 0.0 | 0.1 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.0 | 0.1 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) |
| 0.0 | 0.1 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.2 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 1.2 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 0.3 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.2 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.1 | GO:0035912 | dorsal aorta morphogenesis(GO:0035912) negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.0 | 0.1 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.8 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.2 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.1 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.0 | 0.2 | GO:1901419 | regulation of response to alcohol(GO:1901419) |
| 0.0 | 0.2 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) negative regulation of nuclear division(GO:0051784) |
| 0.0 | 0.6 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 2.7 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.0 | 0.3 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.8 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.5 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.8 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.3 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.1 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.4 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.1 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.2 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.9 | GO:0018279 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.2 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.1 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 1.2 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.0 | 0.1 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 0.3 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.4 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.2 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.1 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.1 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.1 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.0 | 2.3 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.0 | GO:0006524 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) |
| 0.0 | 1.1 | GO:0046323 | glucose import(GO:0046323) |
| 0.0 | 0.8 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.1 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:1904903 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.0 | 0.1 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) cell cycle G2/M phase transition(GO:0044839) |
| 0.0 | 0.1 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.6 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.3 | GO:0006986 | response to unfolded protein(GO:0006986) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:0070695 | FHF complex(GO:0070695) |
| 0.4 | 3.5 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.3 | 1.7 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.3 | 1.6 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) extrinsic component of endosome membrane(GO:0031313) |
| 0.3 | 2.8 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.2 | 1.4 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.2 | 1.8 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.2 | 2.2 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.2 | 1.3 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.2 | 0.8 | GO:1990393 | 3M complex(GO:1990393) |
| 0.1 | 4.3 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 0.2 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 2.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 0.7 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 0.5 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 8.3 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 0.9 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.7 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.1 | 1.1 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.1 | 4.2 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.1 | 0.8 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.2 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.3 | GO:0043219 | lateral loop(GO:0043219) |
| 0.0 | 0.8 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.1 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 0.6 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.2 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.1 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.2 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.1 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.0 | 4.2 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.0 | 0.2 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.1 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.0 | 0.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.0 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 1.4 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.4 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 2.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.3 | GO:0031094 | platelet dense tubular network(GO:0031094) platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.3 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.4 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.0 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.1 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 2.0 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 0.2 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.5 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.4 | 4.4 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.3 | 2.0 | GO:0071253 | connexin binding(GO:0071253) |
| 0.3 | 1.6 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.2 | 0.9 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.2 | 0.7 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.2 | 0.5 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.2 | 1.7 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.2 | 2.6 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.1 | 0.6 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.1 | 0.7 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.1 | 0.4 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 0.3 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.1 | 0.9 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 1.1 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.1 | 0.3 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.1 | 0.5 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 1.4 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 0.4 | GO:0004473 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) oxaloacetate decarboxylase activity(GO:0008948) |
| 0.1 | 2.8 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 0.3 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.1 | 0.6 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.1 | 1.9 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 1.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.3 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.1 | 0.7 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.1 | 0.2 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 0.3 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.5 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 1.0 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.3 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 1.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.8 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.2 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.6 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 1.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.0 | 0.1 | GO:0052827 | inositol pentakisphosphate phosphatase activity(GO:0052827) |
| 0.0 | 1.8 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 2.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.7 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.4 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.2 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 1.0 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 2.2 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.2 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.2 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 1.4 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 0.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.3 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.2 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 1.4 | GO:0004536 | deoxyribonuclease activity(GO:0004536) |
| 0.0 | 0.3 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.1 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.1 | GO:0052812 | phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.2 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.1 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 1.2 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.1 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.1 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 9.3 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.2 | GO:0005521 | lamin binding(GO:0005521) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 3.0 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 2.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 2.7 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 3.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 3.0 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 1.9 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 1.2 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.8 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 1.2 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.7 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.6 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.6 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 2.7 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.5 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.5 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.1 | REACTOME SIGNALING BY NOTCH2 | Genes involved in Signaling by NOTCH2 |
| 0.0 | 1.2 | REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.0 | 0.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.7 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 1.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.3 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 1.2 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.1 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.4 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.2 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.5 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 2.2 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 2.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.9 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 2.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 2.6 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 1.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 2.6 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.3 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.6 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.8 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.4 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.7 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.0 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.3 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |