Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
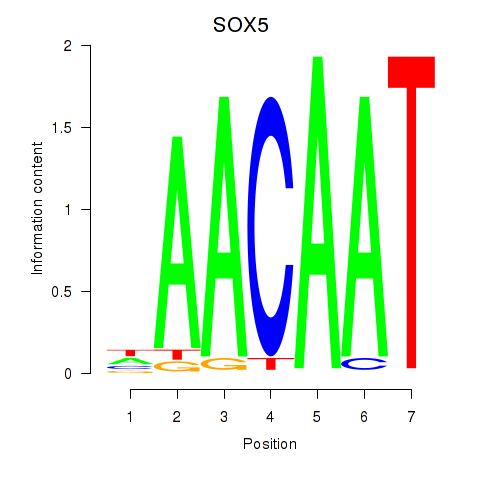
Results for SOX5
Z-value: 1.82
Transcription factors associated with SOX5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX5
|
ENSG00000134532.11 | SOX5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX5 | hg19_v2_chr12_-_24103954_24103972, hg19_v2_chr12_-_23737534_23737550 | 0.37 | 1.6e-01 | Click! |
Activity profile of SOX5 motif
Sorted Z-values of SOX5 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX5
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_6233828 | 4.90 |
ENST00000572068.1 ENST00000261405.5 |
VWF |
von Willebrand factor |
| chr7_-_111846435 | 4.35 |
ENST00000437633.1 ENST00000428084.1 |
DOCK4 |
dedicator of cytokinesis 4 |
| chr6_+_12290586 | 3.97 |
ENST00000379375.5 |
EDN1 |
endothelin 1 |
| chr11_+_46402583 | 3.13 |
ENST00000359803.3 |
MDK |
midkine (neurite growth-promoting factor 2) |
| chr1_-_86043921 | 2.92 |
ENST00000535924.2 |
DDAH1 |
dimethylarginine dimethylaminohydrolase 1 |
| chrY_+_15016013 | 2.86 |
ENST00000360160.4 ENST00000454054.1 |
DDX3Y |
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
| chrX_+_15525426 | 2.84 |
ENST00000342014.6 |
BMX |
BMX non-receptor tyrosine kinase |
| chr11_+_46402744 | 2.78 |
ENST00000533952.1 |
MDK |
midkine (neurite growth-promoting factor 2) |
| chr3_-_99594948 | 2.76 |
ENST00000471562.1 ENST00000495625.2 |
FILIP1L |
filamin A interacting protein 1-like |
| chr3_-_99595037 | 2.58 |
ENST00000383694.2 |
FILIP1L |
filamin A interacting protein 1-like |
| chr2_-_188312971 | 2.57 |
ENST00000410068.1 ENST00000447403.1 ENST00000410102.1 |
CALCRL |
calcitonin receptor-like |
| chr7_-_27205136 | 2.48 |
ENST00000396345.1 ENST00000343483.6 |
HOXA9 |
homeobox A9 |
| chr7_-_131241361 | 2.23 |
ENST00000378555.3 ENST00000322985.9 ENST00000541194.1 ENST00000537928.1 |
PODXL |
podocalyxin-like |
| chr4_-_186732048 | 2.18 |
ENST00000448662.2 ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr4_-_101439148 | 2.14 |
ENST00000511970.1 ENST00000502569.1 ENST00000305864.3 |
EMCN |
endomucin |
| chr4_-_101439242 | 2.11 |
ENST00000296420.4 |
EMCN |
endomucin |
| chr11_+_46402482 | 2.10 |
ENST00000441869.1 |
MDK |
midkine (neurite growth-promoting factor 2) |
| chr3_-_8686479 | 2.09 |
ENST00000544814.1 ENST00000427408.1 |
SSUH2 |
ssu-2 homolog (C. elegans) |
| chrX_-_99891796 | 2.02 |
ENST00000373020.4 |
TSPAN6 |
tetraspanin 6 |
| chr2_-_133427767 | 1.95 |
ENST00000397463.2 |
LYPD1 |
LY6/PLAUR domain containing 1 |
| chr7_-_47621736 | 1.86 |
ENST00000311160.9 |
TNS3 |
tensin 3 |
| chr18_-_52989217 | 1.83 |
ENST00000570287.2 |
TCF4 |
transcription factor 4 |
| chr11_-_89224638 | 1.81 |
ENST00000535633.1 ENST00000263317.4 |
NOX4 |
NADPH oxidase 4 |
| chr20_-_45985172 | 1.76 |
ENST00000536340.1 |
ZMYND8 |
zinc finger, MYND-type containing 8 |
| chr7_-_11871815 | 1.72 |
ENST00000423059.4 |
THSD7A |
thrombospondin, type I, domain containing 7A |
| chr21_-_39870339 | 1.68 |
ENST00000429727.2 ENST00000398905.1 ENST00000398907.1 ENST00000453032.2 ENST00000288319.7 |
ERG |
v-ets avian erythroblastosis virus E26 oncogene homolog |
| chr2_-_160473114 | 1.67 |
ENST00000392783.2 |
BAZ2B |
bromodomain adjacent to zinc finger domain, 2B |
| chr10_-_116444371 | 1.65 |
ENST00000533213.2 ENST00000369252.4 |
ABLIM1 |
actin binding LIM protein 1 |
| chr14_-_89883412 | 1.51 |
ENST00000557258.1 |
FOXN3 |
forkhead box N3 |
| chr6_+_89790490 | 1.50 |
ENST00000336032.3 |
PNRC1 |
proline-rich nuclear receptor coactivator 1 |
| chr18_-_52989525 | 1.49 |
ENST00000457482.3 |
TCF4 |
transcription factor 4 |
| chr12_-_71031185 | 1.46 |
ENST00000548122.1 ENST00000551525.1 ENST00000550358.1 |
PTPRB |
protein tyrosine phosphatase, receptor type, B |
| chr11_+_19799327 | 1.46 |
ENST00000540292.1 |
NAV2 |
neuron navigator 2 |
| chr2_-_192711968 | 1.45 |
ENST00000304141.4 |
SDPR |
serum deprivation response |
| chr6_+_89790459 | 1.44 |
ENST00000369472.1 |
PNRC1 |
proline-rich nuclear receptor coactivator 1 |
| chr12_-_71003568 | 1.43 |
ENST00000547715.1 ENST00000451516.2 ENST00000538708.1 ENST00000550857.1 ENST00000261266.5 |
PTPRB |
protein tyrosine phosphatase, receptor type, B |
| chr18_-_53177984 | 1.41 |
ENST00000543082.1 |
TCF4 |
transcription factor 4 |
| chr1_+_81771806 | 1.37 |
ENST00000370721.1 ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2 |
latrophilin 2 |
| chr11_+_128563948 | 1.35 |
ENST00000534087.2 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
| chr1_-_12677714 | 1.35 |
ENST00000376223.2 |
DHRS3 |
dehydrogenase/reductase (SDR family) member 3 |
| chr5_-_16936340 | 1.33 |
ENST00000507288.1 ENST00000513610.1 |
MYO10 |
myosin X |
| chr6_-_128841503 | 1.33 |
ENST00000368215.3 ENST00000532331.1 ENST00000368213.5 ENST00000368207.3 ENST00000525459.1 ENST00000368210.3 ENST00000368226.4 ENST00000368227.3 |
PTPRK |
protein tyrosine phosphatase, receptor type, K |
| chr11_+_19798964 | 1.27 |
ENST00000527559.2 |
NAV2 |
neuron navigator 2 |
| chr11_+_128563652 | 1.25 |
ENST00000527786.2 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
| chr3_+_173116225 | 1.20 |
ENST00000457714.1 |
NLGN1 |
neuroligin 1 |
| chr12_-_89746173 | 1.20 |
ENST00000308385.6 |
DUSP6 |
dual specificity phosphatase 6 |
| chr20_-_45985464 | 1.16 |
ENST00000458360.2 ENST00000262975.4 |
ZMYND8 |
zinc finger, MYND-type containing 8 |
| chr20_-_45981138 | 1.15 |
ENST00000446994.2 |
ZMYND8 |
zinc finger, MYND-type containing 8 |
| chr14_+_61654271 | 1.14 |
ENST00000555185.1 ENST00000557294.1 ENST00000556778.1 |
PRKCH |
protein kinase C, eta |
| chr14_-_55878538 | 1.11 |
ENST00000247178.5 |
ATG14 |
autophagy related 14 |
| chr11_+_128562372 | 1.11 |
ENST00000344954.6 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
| chr10_-_13523073 | 1.11 |
ENST00000440282.1 |
BEND7 |
BEN domain containing 7 |
| chr14_+_77228532 | 1.07 |
ENST00000167106.4 ENST00000554237.1 |
VASH1 |
vasohibin 1 |
| chr20_-_45985414 | 1.03 |
ENST00000461685.1 ENST00000372023.3 ENST00000540497.1 ENST00000435836.1 ENST00000471951.2 ENST00000352431.2 ENST00000396281.4 ENST00000355972.4 ENST00000360911.3 |
ZMYND8 |
zinc finger, MYND-type containing 8 |
| chr1_+_10271674 | 1.02 |
ENST00000377086.1 |
KIF1B |
kinesin family member 1B |
| chr3_+_25469802 | 1.00 |
ENST00000330688.4 |
RARB |
retinoic acid receptor, beta |
| chr8_-_93115445 | 0.99 |
ENST00000523629.1 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr12_-_71031220 | 0.96 |
ENST00000334414.6 |
PTPRB |
protein tyrosine phosphatase, receptor type, B |
| chr2_-_70475701 | 0.89 |
ENST00000282574.4 |
TIA1 |
TIA1 cytotoxic granule-associated RNA binding protein |
| chr11_-_35441524 | 0.87 |
ENST00000395750.1 ENST00000449068.1 |
SLC1A2 |
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr2_-_70475730 | 0.85 |
ENST00000445587.1 ENST00000433529.2 ENST00000415783.2 |
TIA1 |
TIA1 cytotoxic granule-associated RNA binding protein |
| chrX_-_64196376 | 0.85 |
ENST00000447788.2 |
ZC4H2 |
zinc finger, C4H2 domain containing |
| chr3_-_114343039 | 0.82 |
ENST00000481632.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr2_+_102314161 | 0.82 |
ENST00000425019.1 |
MAP4K4 |
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr11_-_102323489 | 0.80 |
ENST00000361236.3 |
TMEM123 |
transmembrane protein 123 |
| chr7_+_29519486 | 0.80 |
ENST00000409041.4 |
CHN2 |
chimerin 2 |
| chr6_-_152957944 | 0.79 |
ENST00000423061.1 |
SYNE1 |
spectrin repeat containing, nuclear envelope 1 |
| chr10_+_97803151 | 0.78 |
ENST00000403870.3 ENST00000265992.5 ENST00000465148.2 ENST00000534974.1 |
CCNJ |
cyclin J |
| chr12_+_52306113 | 0.77 |
ENST00000547400.1 ENST00000550683.1 ENST00000419526.2 |
ACVRL1 |
activin A receptor type II-like 1 |
| chr3_-_114790179 | 0.77 |
ENST00000462705.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr11_-_102323740 | 0.76 |
ENST00000398136.2 |
TMEM123 |
transmembrane protein 123 |
| chr7_+_134576317 | 0.76 |
ENST00000424922.1 ENST00000495522.1 |
CALD1 |
caldesmon 1 |
| chr11_-_35440796 | 0.75 |
ENST00000278379.3 |
SLC1A2 |
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr2_+_33661382 | 0.75 |
ENST00000402538.3 |
RASGRP3 |
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chrX_+_107683096 | 0.73 |
ENST00000328300.6 ENST00000361603.2 |
COL4A5 |
collagen, type IV, alpha 5 |
| chr15_+_66994561 | 0.73 |
ENST00000288840.5 |
SMAD6 |
SMAD family member 6 |
| chr3_-_129407535 | 0.73 |
ENST00000432054.2 |
TMCC1 |
transmembrane and coiled-coil domain family 1 |
| chr10_+_99079008 | 0.73 |
ENST00000371021.3 |
FRAT1 |
frequently rearranged in advanced T-cell lymphomas |
| chr2_-_160472952 | 0.70 |
ENST00000541068.2 ENST00000355831.2 ENST00000343439.5 ENST00000392782.1 |
BAZ2B |
bromodomain adjacent to zinc finger domain, 2B |
| chr9_+_27109133 | 0.69 |
ENST00000519097.1 ENST00000380036.4 |
TEK |
TEK tyrosine kinase, endothelial |
| chr3_-_11685345 | 0.69 |
ENST00000430365.2 |
VGLL4 |
vestigial like 4 (Drosophila) |
| chr12_-_498620 | 0.68 |
ENST00000399788.2 ENST00000382815.4 |
KDM5A |
lysine (K)-specific demethylase 5A |
| chr16_-_73093597 | 0.67 |
ENST00000397992.5 |
ZFHX3 |
zinc finger homeobox 3 |
| chr3_+_25469724 | 0.67 |
ENST00000437042.2 |
RARB |
retinoic acid receptor, beta |
| chr2_+_86668464 | 0.66 |
ENST00000409064.1 |
KDM3A |
lysine (K)-specific demethylase 3A |
| chr7_+_134576151 | 0.66 |
ENST00000393118.2 |
CALD1 |
caldesmon 1 |
| chr7_+_134551583 | 0.65 |
ENST00000435928.1 |
CALD1 |
caldesmon 1 |
| chr7_+_111846643 | 0.62 |
ENST00000361822.3 |
ZNF277 |
zinc finger protein 277 |
| chr8_-_57123815 | 0.61 |
ENST00000316981.3 ENST00000423799.2 ENST00000429357.2 |
PLAG1 |
pleiomorphic adenoma gene 1 |
| chr17_+_37894570 | 0.60 |
ENST00000394211.3 |
GRB7 |
growth factor receptor-bound protein 7 |
| chr14_-_92413727 | 0.58 |
ENST00000267620.10 |
FBLN5 |
fibulin 5 |
| chr12_+_53443680 | 0.58 |
ENST00000314250.6 ENST00000451358.1 |
TENC1 |
tensin like C1 domain containing phosphatase (tensin 2) |
| chr10_+_104535994 | 0.57 |
ENST00000369889.4 |
WBP1L |
WW domain binding protein 1-like |
| chr20_-_45984401 | 0.57 |
ENST00000311275.7 |
ZMYND8 |
zinc finger, MYND-type containing 8 |
| chr12_-_111926342 | 0.56 |
ENST00000389154.3 |
ATXN2 |
ataxin 2 |
| chr1_+_163038565 | 0.56 |
ENST00000421743.2 |
RGS4 |
regulator of G-protein signaling 4 |
| chr9_-_14314518 | 0.56 |
ENST00000397581.2 |
NFIB |
nuclear factor I/B |
| chr9_-_124989804 | 0.53 |
ENST00000373755.2 ENST00000373754.2 |
LHX6 |
LIM homeobox 6 |
| chr17_+_37894179 | 0.53 |
ENST00000577695.1 ENST00000309156.4 ENST00000309185.3 |
GRB7 |
growth factor receptor-bound protein 7 |
| chr1_-_243418621 | 0.52 |
ENST00000366544.1 ENST00000366543.1 |
CEP170 |
centrosomal protein 170kDa |
| chr18_+_3451646 | 0.52 |
ENST00000345133.5 ENST00000330513.5 ENST00000549546.1 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chr1_+_150337100 | 0.51 |
ENST00000401000.4 |
RPRD2 |
regulation of nuclear pre-mRNA domain containing 2 |
| chr9_-_15510989 | 0.50 |
ENST00000380715.1 ENST00000380716.4 ENST00000380738.4 ENST00000380733.4 |
PSIP1 |
PC4 and SFRS1 interacting protein 1 |
| chr9_-_14314566 | 0.50 |
ENST00000397579.2 |
NFIB |
nuclear factor I/B |
| chr12_+_53443963 | 0.49 |
ENST00000546602.1 ENST00000552570.1 ENST00000549700.1 |
TENC1 |
tensin like C1 domain containing phosphatase (tensin 2) |
| chr2_-_26205340 | 0.49 |
ENST00000264712.3 |
KIF3C |
kinesin family member 3C |
| chr17_+_67498538 | 0.49 |
ENST00000589647.1 |
MAP2K6 |
mitogen-activated protein kinase kinase 6 |
| chr5_+_179159813 | 0.48 |
ENST00000292599.3 |
MAML1 |
mastermind-like 1 (Drosophila) |
| chr22_-_36236265 | 0.46 |
ENST00000414461.2 ENST00000416721.2 ENST00000449924.2 ENST00000262829.7 ENST00000397305.3 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr2_+_30569506 | 0.46 |
ENST00000421976.2 |
AC109642.1 |
AC109642.1 |
| chrX_+_123094369 | 0.44 |
ENST00000455404.1 ENST00000218089.9 |
STAG2 |
stromal antigen 2 |
| chr1_+_150337144 | 0.44 |
ENST00000539519.1 ENST00000369067.3 ENST00000369068.4 |
RPRD2 |
regulation of nuclear pre-mRNA domain containing 2 |
| chr1_-_150241341 | 0.44 |
ENST00000369109.3 ENST00000414276.2 ENST00000236017.5 |
APH1A |
APH1A gamma secretase subunit |
| chrX_-_100872911 | 0.44 |
ENST00000361910.4 ENST00000539247.1 ENST00000538627.1 |
ARMCX6 |
armadillo repeat containing, X-linked 6 |
| chr4_-_76598326 | 0.43 |
ENST00000503660.1 |
G3BP2 |
GTPase activating protein (SH3 domain) binding protein 2 |
| chr9_-_37465396 | 0.42 |
ENST00000307750.4 |
ZBTB5 |
zinc finger and BTB domain containing 5 |
| chr11_+_111807863 | 0.41 |
ENST00000440460.2 |
DIXDC1 |
DIX domain containing 1 |
| chr11_-_35441597 | 0.40 |
ENST00000395753.1 |
SLC1A2 |
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr3_+_189507460 | 0.40 |
ENST00000434928.1 |
TP63 |
tumor protein p63 |
| chr1_+_167298281 | 0.36 |
ENST00000367862.5 |
POU2F1 |
POU class 2 homeobox 1 |
| chr1_-_151119087 | 0.36 |
ENST00000341697.3 ENST00000368914.3 |
SEMA6C |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6C |
| chr6_+_37137939 | 0.36 |
ENST00000373509.5 |
PIM1 |
pim-1 oncogene |
| chr22_+_38093005 | 0.35 |
ENST00000406386.3 |
TRIOBP |
TRIO and F-actin binding protein |
| chrX_-_50557302 | 0.35 |
ENST00000289292.7 |
SHROOM4 |
shroom family member 4 |
| chr1_-_243418344 | 0.34 |
ENST00000366542.1 |
CEP170 |
centrosomal protein 170kDa |
| chr10_+_63661053 | 0.34 |
ENST00000279873.7 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
| chr14_+_23790655 | 0.34 |
ENST00000397276.2 |
PABPN1 |
poly(A) binding protein, nuclear 1 |
| chr2_+_58655461 | 0.33 |
ENST00000429095.1 ENST00000429664.1 ENST00000452840.1 |
AC007092.1 |
long intergenic non-protein coding RNA 1122 |
| chr1_+_147013182 | 0.31 |
ENST00000234739.3 |
BCL9 |
B-cell CLL/lymphoma 9 |
| chr5_-_175964366 | 0.31 |
ENST00000274811.4 |
RNF44 |
ring finger protein 44 |
| chr8_-_33424636 | 0.30 |
ENST00000256257.1 |
RNF122 |
ring finger protein 122 |
| chr6_-_99873145 | 0.30 |
ENST00000369239.5 ENST00000438806.1 |
PNISR |
PNN-interacting serine/arginine-rich protein |
| chr7_+_29519662 | 0.29 |
ENST00000424025.2 ENST00000439711.2 ENST00000421775.2 |
CHN2 |
chimerin 2 |
| chr18_+_3451584 | 0.29 |
ENST00000551541.1 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chr16_+_3704822 | 0.28 |
ENST00000414110.2 |
DNASE1 |
deoxyribonuclease I |
| chr4_+_110749143 | 0.28 |
ENST00000317735.4 |
RRH |
retinal pigment epithelium-derived rhodopsin homolog |
| chr6_+_119215308 | 0.28 |
ENST00000229595.5 |
ASF1A |
anti-silencing function 1A histone chaperone |
| chr7_-_32931387 | 0.28 |
ENST00000304056.4 |
KBTBD2 |
kelch repeat and BTB (POZ) domain containing 2 |
| chr8_-_72274467 | 0.28 |
ENST00000340726.3 |
EYA1 |
eyes absent homolog 1 (Drosophila) |
| chr20_+_32150140 | 0.27 |
ENST00000344201.3 ENST00000346541.3 ENST00000397800.1 ENST00000397798.2 ENST00000492345.1 |
CBFA2T2 |
core-binding factor, runt domain, alpha subunit 2; translocated to, 2 |
| chr1_+_33116743 | 0.25 |
ENST00000414241.3 ENST00000373493.5 |
RBBP4 |
retinoblastoma binding protein 4 |
| chr6_+_56954867 | 0.24 |
ENST00000370708.4 ENST00000370702.1 |
ZNF451 |
zinc finger protein 451 |
| chr4_+_160188889 | 0.24 |
ENST00000264431.4 |
RAPGEF2 |
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr11_-_82708435 | 0.23 |
ENST00000525117.1 ENST00000532548.1 |
RAB30 |
RAB30, member RAS oncogene family |
| chr12_+_49761273 | 0.23 |
ENST00000551540.1 ENST00000552918.1 ENST00000548777.1 ENST00000547865.1 ENST00000552171.1 |
SPATS2 |
spermatogenesis associated, serine-rich 2 |
| chr11_-_82708519 | 0.22 |
ENST00000534301.1 |
RAB30 |
RAB30, member RAS oncogene family |
| chr15_-_37391507 | 0.22 |
ENST00000557796.2 ENST00000397620.2 |
MEIS2 |
Meis homeobox 2 |
| chr3_-_48754599 | 0.22 |
ENST00000413654.1 ENST00000454335.1 ENST00000440424.1 ENST00000449610.1 ENST00000443964.1 ENST00000417896.1 ENST00000413298.1 ENST00000449563.1 ENST00000443853.1 ENST00000437427.1 ENST00000446860.1 ENST00000412850.1 ENST00000424035.1 ENST00000340879.4 ENST00000431721.2 ENST00000434860.1 ENST00000328631.5 ENST00000432678.2 |
IP6K2 |
inositol hexakisphosphate kinase 2 |
| chr15_-_37391614 | 0.22 |
ENST00000219869.9 |
MEIS2 |
Meis homeobox 2 |
| chr4_-_2264015 | 0.22 |
ENST00000337190.2 |
MXD4 |
MAX dimerization protein 4 |
| chrX_-_50557014 | 0.22 |
ENST00000376020.2 |
SHROOM4 |
shroom family member 4 |
| chr1_+_15272271 | 0.22 |
ENST00000400797.3 |
KAZN |
kazrin, periplakin interacting protein |
| chr6_+_126240442 | 0.21 |
ENST00000448104.1 ENST00000438495.1 ENST00000444128.1 |
NCOA7 |
nuclear receptor coactivator 7 |
| chr9_+_27109392 | 0.20 |
ENST00000406359.4 |
TEK |
TEK tyrosine kinase, endothelial |
| chrX_+_123094672 | 0.20 |
ENST00000354548.5 ENST00000458700.1 |
STAG2 |
stromal antigen 2 |
| chr3_-_52486841 | 0.20 |
ENST00000496590.1 |
TNNC1 |
troponin C type 1 (slow) |
| chr12_-_91451758 | 0.20 |
ENST00000266719.3 |
KERA |
keratocan |
| chr1_-_53018654 | 0.19 |
ENST00000257177.4 ENST00000355809.4 ENST00000528642.1 ENST00000470626.1 ENST00000371544.3 |
ZCCHC11 |
zinc finger, CCHC domain containing 11 |
| chr4_+_41614720 | 0.19 |
ENST00000509277.1 |
LIMCH1 |
LIM and calponin homology domains 1 |
| chr12_+_49761224 | 0.19 |
ENST00000553127.1 ENST00000321898.6 |
SPATS2 |
spermatogenesis associated, serine-rich 2 |
| chr1_+_228337553 | 0.19 |
ENST00000366714.2 |
GJC2 |
gap junction protein, gamma 2, 47kDa |
| chr12_+_52445191 | 0.19 |
ENST00000243050.1 ENST00000394825.1 ENST00000550763.1 ENST00000394824.2 ENST00000548232.1 ENST00000562373.1 |
NR4A1 |
nuclear receptor subfamily 4, group A, member 1 |
| chr1_-_217250231 | 0.18 |
ENST00000493748.1 ENST00000463665.1 |
ESRRG |
estrogen-related receptor gamma |
| chr12_-_51477333 | 0.18 |
ENST00000228515.1 ENST00000548206.1 ENST00000546935.1 ENST00000548981.1 |
CSRNP2 |
cysteine-serine-rich nuclear protein 2 |
| chr15_-_86338100 | 0.18 |
ENST00000536947.1 |
KLHL25 |
kelch-like family member 25 |
| chr4_+_41614909 | 0.18 |
ENST00000509454.1 ENST00000396595.3 ENST00000381753.4 |
LIMCH1 |
LIM and calponin homology domains 1 |
| chr1_-_40367530 | 0.18 |
ENST00000372816.2 ENST00000372815.1 |
MYCL |
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chr12_-_122985067 | 0.17 |
ENST00000540586.1 ENST00000543897.1 |
ZCCHC8 |
zinc finger, CCHC domain containing 8 |
| chr6_-_79944336 | 0.17 |
ENST00000344726.5 ENST00000275036.7 |
HMGN3 |
high mobility group nucleosomal binding domain 3 |
| chr12_-_122985494 | 0.17 |
ENST00000336229.4 |
ZCCHC8 |
zinc finger, CCHC domain containing 8 |
| chrX_+_107288197 | 0.17 |
ENST00000415430.3 |
VSIG1 |
V-set and immunoglobulin domain containing 1 |
| chr14_+_23790690 | 0.16 |
ENST00000556821.1 |
PABPN1 |
poly(A) binding protein, nuclear 1 |
| chr9_-_123239632 | 0.16 |
ENST00000416449.1 |
CDK5RAP2 |
CDK5 regulatory subunit associated protein 2 |
| chr10_+_70480963 | 0.15 |
ENST00000265872.6 ENST00000535016.1 ENST00000538031.1 ENST00000543719.1 ENST00000539539.1 ENST00000543225.1 ENST00000536012.1 ENST00000494903.2 |
CCAR1 |
cell division cycle and apoptosis regulator 1 |
| chr16_+_31085714 | 0.14 |
ENST00000300850.5 ENST00000564189.1 ENST00000428260.1 |
ZNF646 |
zinc finger protein 646 |
| chr1_-_8000872 | 0.14 |
ENST00000377507.3 |
TNFRSF9 |
tumor necrosis factor receptor superfamily, member 9 |
| chr15_+_75074410 | 0.14 |
ENST00000439220.2 |
CSK |
c-src tyrosine kinase |
| chr2_-_208989225 | 0.13 |
ENST00000264376.4 |
CRYGD |
crystallin, gamma D |
| chrX_+_107288239 | 0.13 |
ENST00000217957.5 |
VSIG1 |
V-set and immunoglobulin domain containing 1 |
| chr17_-_39041479 | 0.13 |
ENST00000167588.3 |
KRT20 |
keratin 20 |
| chr10_-_99094458 | 0.13 |
ENST00000371019.2 |
FRAT2 |
frequently rearranged in advanced T-cell lymphomas 2 |
| chr5_-_147162078 | 0.13 |
ENST00000507386.1 |
JAKMIP2 |
janus kinase and microtubule interacting protein 2 |
| chr4_+_170581213 | 0.12 |
ENST00000507875.1 |
CLCN3 |
chloride channel, voltage-sensitive 3 |
| chr3_+_89156799 | 0.12 |
ENST00000452448.2 ENST00000494014.1 |
EPHA3 |
EPH receptor A3 |
| chr15_-_86338134 | 0.12 |
ENST00000337975.5 |
KLHL25 |
kelch-like family member 25 |
| chr11_-_115375107 | 0.11 |
ENST00000545380.1 ENST00000452722.3 ENST00000537058.1 ENST00000536727.1 ENST00000542447.2 ENST00000331581.6 |
CADM1 |
cell adhesion molecule 1 |
| chr8_-_72274095 | 0.11 |
ENST00000303824.7 |
EYA1 |
eyes absent homolog 1 (Drosophila) |
| chr2_-_55277654 | 0.11 |
ENST00000337526.6 ENST00000317610.7 ENST00000357732.4 |
RTN4 |
reticulon 4 |
| chr14_+_22748980 | 0.11 |
ENST00000390465.2 |
TRAV38-2DV8 |
T cell receptor alpha variable 38-2/delta variable 8 |
| chr19_+_38397839 | 0.10 |
ENST00000222345.6 |
SIPA1L3 |
signal-induced proliferation-associated 1 like 3 |
| chr1_-_167883327 | 0.10 |
ENST00000476818.2 ENST00000367851.4 ENST00000367848.1 |
ADCY10 |
adenylate cyclase 10 (soluble) |
| chr2_-_86422523 | 0.09 |
ENST00000442664.2 ENST00000409051.2 ENST00000449247.2 |
IMMT |
inner membrane protein, mitochondrial |
| chr5_-_134871639 | 0.09 |
ENST00000314744.4 |
NEUROG1 |
neurogenin 1 |
| chr1_-_28177255 | 0.09 |
ENST00000601459.1 |
AL109927.1 |
HCG2032222; PRO2047; Uncharacterized protein |
| chr15_-_34610962 | 0.07 |
ENST00000290209.5 |
SLC12A6 |
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr1_-_226926864 | 0.07 |
ENST00000429204.1 ENST00000366784.1 |
ITPKB |
inositol-trisphosphate 3-kinase B |
| chrX_+_70503433 | 0.07 |
ENST00000276079.8 ENST00000373856.3 ENST00000373841.1 ENST00000420903.1 |
NONO |
non-POU domain containing, octamer-binding |
| chr2_-_55277692 | 0.07 |
ENST00000394611.2 |
RTN4 |
reticulon 4 |
| chr9_-_130712995 | 0.07 |
ENST00000373084.4 |
FAM102A |
family with sequence similarity 102, member A |
| chr15_+_75074385 | 0.06 |
ENST00000220003.9 |
CSK |
c-src tyrosine kinase |
Gene Ontology Analysis
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 8.0 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 4.2 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 4.6 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 1.3 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 6.0 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.2 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.5 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.1 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 2.0 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 1.1 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 3.2 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 1.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.6 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.8 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.2 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.8 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 3.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.6 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.0 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.4 | 4.9 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.4 | 1.1 | GO:1903349 | omegasome membrane(GO:1903349) |
| 0.3 | 2.2 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.2 | 1.7 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.2 | 2.7 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 2.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 2.5 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 4.4 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 1.8 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 5.7 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 0.5 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 1.1 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 1.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 0.7 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.1 | 0.8 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 0.3 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 2.0 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 0.8 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.4 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 3.9 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.5 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.2 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 6.4 | GO:0036464 | cytoplasmic ribonucleoprotein granule(GO:0036464) |
| 0.0 | 0.5 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 1.5 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.0 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 5.0 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 1.6 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.7 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 2.8 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.9 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.0 | GO:0020003 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.2 | GO:0005922 | connexon complex(GO:0005922) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.9 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 4.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 1.7 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 0.9 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 0.5 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 1.3 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.5 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.6 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 1.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 2.6 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 1.1 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 1.1 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.4 | REACTOME SIGNALING BY NOTCH2 | Genes involved in Signaling by NOTCH2 |
| 0.0 | 2.0 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.4 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.5 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 3.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 3.8 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.8 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 1.4 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 2.6 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.3 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.0 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.6 | 2.6 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.6 | 2.9 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.4 | 4.7 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.3 | 2.0 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.3 | 2.0 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.2 | 1.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.2 | 0.8 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.2 | 1.3 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.2 | 5.2 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.2 | 1.8 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.2 | 0.7 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.2 | 4.9 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.1 | 1.3 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.1 | 5.7 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 0.6 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.1 | 0.8 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 1.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 2.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 1.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 1.4 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.1 | 2.1 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.3 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.1 | 0.2 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.1 | 4.4 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 0.3 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 9.9 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.2 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.8 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 1.3 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.5 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.0 | 0.7 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 2.9 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.2 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 3.8 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 0.4 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 2.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.2 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.0 | 0.8 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.8 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.3 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.3 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 2.2 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.4 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 1.0 | GO:1990939 | ATP-dependent microtubule motor activity(GO:1990939) |
| 0.0 | 0.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.2 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.4 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.5 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.6 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.0 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 8.0 | GO:0030421 | defecation(GO:0030421) |
| 1.3 | 4.0 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.9 | 5.7 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.9 | 4.4 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.7 | 2.7 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.6 | 2.2 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.4 | 1.2 | GO:0099543 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) |
| 0.4 | 2.9 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.4 | 1.1 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.3 | 1.0 | GO:1904647 | response to rotenone(GO:1904647) |
| 0.3 | 1.7 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.3 | 1.2 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 0.3 | 2.8 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.3 | 2.0 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.2 | 2.6 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.2 | 2.9 | GO:1900038 | negative regulation of cellular response to hypoxia(GO:1900038) |
| 0.2 | 1.1 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.2 | 0.7 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.2 | 2.5 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.2 | 1.7 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.2 | 3.7 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.2 | 0.7 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 0.2 | 2.0 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.2 | 0.5 | GO:1904247 | regulation of polynucleotide adenylyltransferase activity(GO:1904245) positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.2 | 0.7 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.2 | 0.5 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.2 | 0.5 | GO:0060926 | atrioventricular node development(GO:0003162) cardiac pacemaker cell differentiation(GO:0060920) cardiac pacemaker cell development(GO:0060926) |
| 0.2 | 3.7 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.2 | 1.1 | GO:0021740 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 | 1.1 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 | 1.3 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 | 0.9 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.1 | 0.2 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.1 | 1.6 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 1.0 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.1 | 1.3 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 1.3 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 | 0.8 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.1 | 0.2 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) |
| 0.1 | 1.6 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.1 | 2.1 | GO:1904376 | negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.1 | 0.8 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 0.5 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 0.6 | GO:0003183 | mitral valve morphogenesis(GO:0003183) |
| 0.1 | 0.4 | GO:0030047 | actin modification(GO:0030047) |
| 0.1 | 0.4 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.1 | 0.6 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 | 2.9 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 0.7 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 | 0.4 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.1 | 0.5 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.5 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.5 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 | 0.4 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 | 0.6 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.1 | 1.8 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.2 | GO:0032972 | diaphragm contraction(GO:0002086) regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 0.6 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.0 | 1.9 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.2 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.0 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.0 | 1.6 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.9 | GO:1904886 | beta-catenin destruction complex disassembly(GO:1904886) |
| 0.0 | 3.5 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.0 | 0.6 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.8 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 0.4 | GO:0007220 | Notch receptor processing(GO:0007220) membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:2000980 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.2 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 1.0 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 1.7 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.3 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 4.5 | GO:0065004 | protein-DNA complex assembly(GO:0065004) |
| 0.0 | 0.2 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.0 | 1.3 | GO:0009301 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.9 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.1 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.2 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.1 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.3 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.0 | GO:0003415 | chondrocyte hypertrophy(GO:0003415) |
| 0.0 | 0.4 | GO:2000772 | regulation of cellular senescence(GO:2000772) |
| 0.0 | 0.5 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.3 | GO:0016577 | histone demethylation(GO:0016577) |