Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
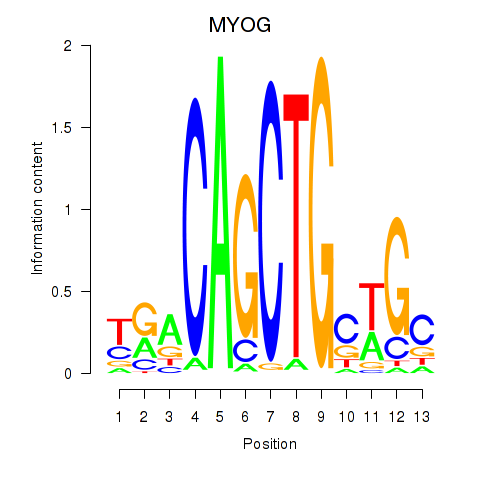
Results for TCF3_MYOG
Z-value: 1.53
Transcription factors associated with TCF3_MYOG
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TCF3
|
ENSG00000071564.10 | TCF3 |
|
MYOG
|
ENSG00000122180.4 | MYOG |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MYOG | hg19_v2_chr1_-_203055129_203055164 | 0.75 | 9.0e-04 | Click! |
| TCF3 | hg19_v2_chr19_-_1650666_1650744 | 0.18 | 5.1e-01 | Click! |
Activity profile of TCF3_MYOG motif
Sorted Z-values of TCF3_MYOG motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TCF3_MYOG
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_45286706 | 6.48 |
ENST00000393450.1 ENST00000572303.1 |
MYL4 |
myosin, light chain 4, alkali; atrial, embryonic |
| chr17_+_45286387 | 6.16 |
ENST00000572316.1 ENST00000354968.1 ENST00000576874.1 ENST00000536623.2 |
MYL4 |
myosin, light chain 4, alkali; atrial, embryonic |
| chr9_-_35689900 | 5.32 |
ENST00000378300.5 ENST00000329305.2 ENST00000360958.2 |
TPM2 |
tropomyosin 2 (beta) |
| chr6_+_44184653 | 4.10 |
ENST00000573382.2 ENST00000576476.1 |
RP1-302G2.5 |
RP1-302G2.5 |
| chr1_-_201391149 | 3.99 |
ENST00000555948.1 ENST00000556362.1 |
TNNI1 |
troponin I type 1 (skeletal, slow) |
| chr21_+_30502806 | 3.94 |
ENST00000399928.1 ENST00000399926.1 |
MAP3K7CL |
MAP3K7 C-terminal like |
| chr16_+_30383613 | 3.77 |
ENST00000568749.1 |
MYLPF |
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr3_-_87040233 | 3.65 |
ENST00000398399.2 |
VGLL3 |
vestigial like 3 (Drosophila) |
| chr2_-_152590946 | 3.41 |
ENST00000172853.10 |
NEB |
nebulin |
| chr8_+_98788003 | 3.41 |
ENST00000521545.2 |
LAPTM4B |
lysosomal protein transmembrane 4 beta |
| chr2_+_220283091 | 3.19 |
ENST00000373960.3 |
DES |
desmin |
| chr2_+_170366203 | 3.09 |
ENST00000284669.1 |
KLHL41 |
kelch-like family member 41 |
| chr2_-_37899323 | 2.84 |
ENST00000295324.3 ENST00000457889.1 |
CDC42EP3 |
CDC42 effector protein (Rho GTPase binding) 3 |
| chrX_-_11445856 | 2.77 |
ENST00000380736.1 |
ARHGAP6 |
Rho GTPase activating protein 6 |
| chr6_+_129204337 | 2.71 |
ENST00000421865.2 |
LAMA2 |
laminin, alpha 2 |
| chr1_-_33168336 | 2.52 |
ENST00000373484.3 |
SYNC |
syncoilin, intermediate filament protein |
| chr3_+_69788576 | 2.51 |
ENST00000352241.4 ENST00000448226.2 |
MITF |
microphthalmia-associated transcription factor |
| chr17_-_10452929 | 2.46 |
ENST00000532183.2 ENST00000397183.2 ENST00000420805.1 |
MYH2 |
myosin, heavy chain 2, skeletal muscle, adult |
| chr1_+_172422026 | 2.45 |
ENST00000367725.4 |
C1orf105 |
chromosome 1 open reading frame 105 |
| chr8_-_41522779 | 2.29 |
ENST00000522231.1 ENST00000314214.8 ENST00000348036.4 ENST00000457297.1 ENST00000522543.1 |
ANK1 |
ankyrin 1, erythrocytic |
| chr7_+_102553430 | 2.23 |
ENST00000339431.4 ENST00000249377.4 |
LRRC17 |
leucine rich repeat containing 17 |
| chr15_-_48937982 | 2.18 |
ENST00000316623.5 |
FBN1 |
fibrillin 1 |
| chr11_-_47470591 | 2.17 |
ENST00000524487.1 |
RAPSN |
receptor-associated protein of the synapse |
| chr3_+_35681081 | 2.13 |
ENST00000428373.1 |
ARPP21 |
cAMP-regulated phosphoprotein, 21kDa |
| chr1_-_201346761 | 2.09 |
ENST00000455702.1 ENST00000422165.1 ENST00000367318.5 ENST00000367320.2 ENST00000438742.1 ENST00000412633.1 ENST00000458432.2 ENST00000421663.2 ENST00000367322.1 ENST00000509001.1 |
TNNT2 |
troponin T type 2 (cardiac) |
| chr11_-_47470703 | 2.06 |
ENST00000298854.2 |
RAPSN |
receptor-associated protein of the synapse |
| chr4_-_186456652 | 2.03 |
ENST00000284767.5 ENST00000284770.5 |
PDLIM3 |
PDZ and LIM domain 3 |
| chr4_-_186456766 | 2.02 |
ENST00000284771.6 |
PDLIM3 |
PDZ and LIM domain 3 |
| chrX_+_70521584 | 2.00 |
ENST00000373829.3 ENST00000538820.1 |
ITGB1BP2 |
integrin beta 1 binding protein (melusin) 2 |
| chr16_+_30387141 | 1.99 |
ENST00000566955.1 |
MYLPF |
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr7_+_98246588 | 1.97 |
ENST00000265634.3 |
NPTX2 |
neuronal pentraxin II |
| chr19_-_45826125 | 1.93 |
ENST00000221476.3 |
CKM |
creatine kinase, muscle |
| chr3_-_52486841 | 1.87 |
ENST00000496590.1 |
TNNC1 |
troponin C type 1 (slow) |
| chr3_+_159570722 | 1.85 |
ENST00000482804.1 |
SCHIP1 |
schwannomin interacting protein 1 |
| chr11_-_64014379 | 1.85 |
ENST00000309318.3 |
PPP1R14B |
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr6_-_31697255 | 1.84 |
ENST00000436437.1 |
DDAH2 |
dimethylarginine dimethylaminohydrolase 2 |
| chr16_-_31076332 | 1.81 |
ENST00000539836.3 ENST00000535577.1 ENST00000442862.2 |
ZNF668 |
zinc finger protein 668 |
| chr11_-_47470682 | 1.80 |
ENST00000529341.1 ENST00000352508.3 |
RAPSN |
receptor-associated protein of the synapse |
| chr16_+_30386098 | 1.79 |
ENST00000322861.7 |
MYLPF |
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr11_+_111783450 | 1.72 |
ENST00000537382.1 |
HSPB2 |
Homo sapiens heat shock 27kDa protein 2 (HSPB2), mRNA. |
| chr7_+_116166331 | 1.69 |
ENST00000393468.1 ENST00000393467.1 |
CAV1 |
caveolin 1, caveolae protein, 22kDa |
| chr9_-_79307096 | 1.69 |
ENST00000376717.2 ENST00000223609.6 ENST00000443509.2 |
PRUNE2 |
prune homolog 2 (Drosophila) |
| chr22_+_38071615 | 1.67 |
ENST00000215909.5 |
LGALS1 |
lectin, galactoside-binding, soluble, 1 |
| chr11_-_1643368 | 1.67 |
ENST00000399682.1 |
KRTAP5-4 |
keratin associated protein 5-4 |
| chrX_-_107018969 | 1.66 |
ENST00000372383.4 |
TSC22D3 |
TSC22 domain family, member 3 |
| chr1_-_193155729 | 1.63 |
ENST00000367434.4 |
B3GALT2 |
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chr7_+_116165754 | 1.61 |
ENST00000405348.1 |
CAV1 |
caveolin 1, caveolae protein, 22kDa |
| chr3_+_8775466 | 1.60 |
ENST00000343849.2 ENST00000397368.2 |
CAV3 |
caveolin 3 |
| chr16_+_7382745 | 1.58 |
ENST00000436368.2 ENST00000311745.5 ENST00000355637.4 ENST00000340209.4 |
RBFOX1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr13_+_102104980 | 1.58 |
ENST00000545560.2 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr4_+_169418195 | 1.57 |
ENST00000261509.6 ENST00000335742.7 |
PALLD |
palladin, cytoskeletal associated protein |
| chr7_+_128470431 | 1.57 |
ENST00000325888.8 ENST00000346177.6 |
FLNC |
filamin C, gamma |
| chr11_-_85779786 | 1.55 |
ENST00000356360.5 |
PICALM |
phosphatidylinositol binding clathrin assembly protein |
| chr19_-_46285646 | 1.54 |
ENST00000458663.2 |
DMPK |
dystrophia myotonica-protein kinase |
| chr13_+_102104952 | 1.54 |
ENST00000376180.3 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr8_-_49834299 | 1.52 |
ENST00000396822.1 |
SNAI2 |
snail family zinc finger 2 |
| chr6_+_39760783 | 1.51 |
ENST00000398904.2 ENST00000538976.1 |
DAAM2 |
dishevelled associated activator of morphogenesis 2 |
| chr11_-_63933504 | 1.48 |
ENST00000255681.6 |
MACROD1 |
MACRO domain containing 1 |
| chr12_+_56114151 | 1.47 |
ENST00000547072.1 ENST00000552930.1 ENST00000257895.5 |
RDH5 |
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr17_+_65040678 | 1.46 |
ENST00000226021.3 |
CACNG1 |
calcium channel, voltage-dependent, gamma subunit 1 |
| chr1_+_25071848 | 1.46 |
ENST00000374379.4 |
CLIC4 |
chloride intracellular channel 4 |
| chr8_-_49833978 | 1.45 |
ENST00000020945.1 |
SNAI2 |
snail family zinc finger 2 |
| chr12_+_56114189 | 1.44 |
ENST00000548082.1 |
RDH5 |
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr2_-_1748214 | 1.42 |
ENST00000433670.1 ENST00000425171.1 ENST00000252804.4 |
PXDN |
peroxidasin homolog (Drosophila) |
| chr2_+_24714729 | 1.40 |
ENST00000406961.1 ENST00000405141.1 |
NCOA1 |
nuclear receptor coactivator 1 |
| chr7_-_150675372 | 1.39 |
ENST00000262186.5 |
KCNH2 |
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| chr11_-_111783919 | 1.39 |
ENST00000531198.1 ENST00000533879.1 |
CRYAB |
crystallin, alpha B |
| chr12_-_110434021 | 1.38 |
ENST00000355312.3 ENST00000551209.1 ENST00000550186.1 |
GIT2 |
G protein-coupled receptor kinase interacting ArfGAP 2 |
| chr9_-_35685452 | 1.37 |
ENST00000607559.1 |
TPM2 |
tropomyosin 2 (beta) |
| chr11_+_86511569 | 1.31 |
ENST00000441050.1 |
PRSS23 |
protease, serine, 23 |
| chr17_-_15168624 | 1.29 |
ENST00000312280.3 ENST00000494511.1 ENST00000580584.1 |
PMP22 |
peripheral myelin protein 22 |
| chr19_-_46285736 | 1.28 |
ENST00000291270.4 ENST00000447742.2 ENST00000354227.5 |
DMPK |
dystrophia myotonica-protein kinase |
| chr12_-_56122426 | 1.27 |
ENST00000551173.1 |
CD63 |
CD63 molecule |
| chr7_-_45960850 | 1.27 |
ENST00000381083.4 ENST00000381086.5 ENST00000275521.6 |
IGFBP3 |
insulin-like growth factor binding protein 3 |
| chr1_-_85156216 | 1.26 |
ENST00000342203.3 ENST00000370612.4 |
SSX2IP |
synovial sarcoma, X breakpoint 2 interacting protein |
| chr8_-_41522719 | 1.23 |
ENST00000335651.6 |
ANK1 |
ankyrin 1, erythrocytic |
| chr10_-_79398250 | 1.22 |
ENST00000286627.5 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr11_-_10829851 | 1.22 |
ENST00000532082.1 |
EIF4G2 |
eukaryotic translation initiation factor 4 gamma, 2 |
| chr1_-_85156090 | 1.20 |
ENST00000605755.1 ENST00000437941.2 |
SSX2IP |
synovial sarcoma, X breakpoint 2 interacting protein |
| chrX_+_135229600 | 1.20 |
ENST00000370690.3 |
FHL1 |
four and a half LIM domains 1 |
| chr4_+_169418255 | 1.18 |
ENST00000505667.1 ENST00000511948.1 |
PALLD |
palladin, cytoskeletal associated protein |
| chr2_-_190044480 | 1.18 |
ENST00000374866.3 |
COL5A2 |
collagen, type V, alpha 2 |
| chr11_-_10830463 | 1.17 |
ENST00000527419.1 ENST00000530211.1 ENST00000530702.1 ENST00000524932.1 ENST00000532570.1 |
EIF4G2 |
eukaryotic translation initiation factor 4 gamma, 2 |
| chr10_-_92681033 | 1.16 |
ENST00000371697.3 |
ANKRD1 |
ankyrin repeat domain 1 (cardiac muscle) |
| chr17_+_52978107 | 1.15 |
ENST00000445275.2 |
TOM1L1 |
target of myb1 (chicken)-like 1 |
| chrX_+_80457442 | 1.13 |
ENST00000373212.5 |
SH3BGRL |
SH3 domain binding glutamic acid-rich protein like |
| chr4_-_41216492 | 1.11 |
ENST00000503503.1 ENST00000509446.1 ENST00000503264.1 ENST00000508707.1 ENST00000508593.1 |
APBB2 |
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr17_+_68165657 | 1.07 |
ENST00000243457.3 |
KCNJ2 |
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr11_-_85779971 | 1.06 |
ENST00000393346.3 |
PICALM |
phosphatidylinositol binding clathrin assembly protein |
| chr11_+_1942580 | 1.06 |
ENST00000381558.1 |
TNNT3 |
troponin T type 3 (skeletal, fast) |
| chr16_-_89043377 | 1.05 |
ENST00000436887.2 ENST00000448839.1 ENST00000360302.2 |
CBFA2T3 |
core-binding factor, runt domain, alpha subunit 2; translocated to, 3 |
| chr7_+_12727250 | 1.04 |
ENST00000404894.1 |
ARL4A |
ADP-ribosylation factor-like 4A |
| chr12_-_56122761 | 1.03 |
ENST00000552164.1 ENST00000420846.3 ENST00000257857.4 |
CD63 |
CD63 molecule |
| chr4_-_41216473 | 1.01 |
ENST00000513140.1 |
APBB2 |
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr2_+_241375069 | 1.01 |
ENST00000264039.2 |
GPC1 |
glypican 1 |
| chr14_+_90863327 | 1.00 |
ENST00000356978.4 |
CALM1 |
calmodulin 1 (phosphorylase kinase, delta) |
| chr7_+_12726474 | 1.00 |
ENST00000396662.1 ENST00000356797.3 ENST00000396664.2 |
ARL4A |
ADP-ribosylation factor-like 4A |
| chr4_-_41216619 | 0.99 |
ENST00000508676.1 ENST00000506352.1 ENST00000295974.8 |
APBB2 |
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr12_+_26274917 | 0.99 |
ENST00000538142.1 |
SSPN |
sarcospan |
| chr9_-_13165457 | 0.98 |
ENST00000542239.1 ENST00000538841.1 ENST00000433359.2 |
MPDZ |
multiple PDZ domain protein |
| chr18_-_52989525 | 0.97 |
ENST00000457482.3 |
TCF4 |
transcription factor 4 |
| chr1_-_84464780 | 0.96 |
ENST00000260505.8 |
TTLL7 |
tubulin tyrosine ligase-like family, member 7 |
| chr15_-_56209306 | 0.96 |
ENST00000506154.1 ENST00000338963.2 ENST00000508342.1 |
NEDD4 |
neural precursor cell expressed, developmentally down-regulated 4, E3 ubiquitin protein ligase |
| chr13_-_36705425 | 0.96 |
ENST00000255448.4 ENST00000360631.3 ENST00000379892.4 |
DCLK1 |
doublecortin-like kinase 1 |
| chr6_-_139695757 | 0.95 |
ENST00000367651.2 |
CITED2 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
| chr19_-_10121144 | 0.95 |
ENST00000264828.3 |
COL5A3 |
collagen, type V, alpha 3 |
| chr1_-_85155939 | 0.95 |
ENST00000603677.1 |
SSX2IP |
synovial sarcoma, X breakpoint 2 interacting protein |
| chr1_-_94079648 | 0.93 |
ENST00000370247.3 |
BCAR3 |
breast cancer anti-estrogen resistance 3 |
| chr8_-_38008783 | 0.93 |
ENST00000276449.4 |
STAR |
steroidogenic acute regulatory protein |
| chr8_+_38758737 | 0.93 |
ENST00000521746.1 ENST00000420274.1 |
PLEKHA2 |
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 2 |
| chr1_+_89990431 | 0.92 |
ENST00000330947.2 ENST00000358200.4 |
LRRC8B |
leucine rich repeat containing 8 family, member B |
| chr8_+_70404996 | 0.89 |
ENST00000402687.4 ENST00000419716.3 |
SULF1 |
sulfatase 1 |
| chr17_+_52978156 | 0.89 |
ENST00000348161.4 |
TOM1L1 |
target of myb1 (chicken)-like 1 |
| chr6_-_24911195 | 0.89 |
ENST00000259698.4 |
FAM65B |
family with sequence similarity 65, member B |
| chrX_+_135229731 | 0.88 |
ENST00000420362.1 |
FHL1 |
four and a half LIM domains 1 |
| chr22_-_22292934 | 0.87 |
ENST00000538191.1 ENST00000424647.1 ENST00000407142.1 |
PPM1F |
protein phosphatase, Mg2+/Mn2+ dependent, 1F |
| chrX_-_13835147 | 0.86 |
ENST00000493677.1 ENST00000355135.2 |
GPM6B |
glycoprotein M6B |
| chr2_-_16847084 | 0.86 |
ENST00000406434.1 ENST00000381323.3 |
FAM49A |
family with sequence similarity 49, member A |
| chr11_-_66084508 | 0.86 |
ENST00000311330.3 |
CD248 |
CD248 molecule, endosialin |
| chr12_+_79258444 | 0.86 |
ENST00000261205.4 |
SYT1 |
synaptotagmin I |
| chr5_+_34656569 | 0.83 |
ENST00000428746.2 |
RAI14 |
retinoic acid induced 14 |
| chr12_-_90102594 | 0.83 |
ENST00000428670.3 |
ATP2B1 |
ATPase, Ca++ transporting, plasma membrane 1 |
| chr11_-_6341844 | 0.81 |
ENST00000303927.3 |
PRKCDBP |
protein kinase C, delta binding protein |
| chr3_-_114866084 | 0.81 |
ENST00000357258.3 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr16_-_31076273 | 0.80 |
ENST00000426488.2 |
ZNF668 |
zinc finger protein 668 |
| chr3_-_178790057 | 0.80 |
ENST00000311417.2 |
ZMAT3 |
zinc finger, matrin-type 3 |
| chr8_-_101963482 | 0.80 |
ENST00000419477.2 |
YWHAZ |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr3_+_158787041 | 0.79 |
ENST00000471575.1 ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1 |
IQCJ-SCHIP1 readthrough |
| chr1_-_85930823 | 0.79 |
ENST00000284031.8 ENST00000539042.1 |
DDAH1 |
dimethylarginine dimethylaminohydrolase 1 |
| chr6_-_137113604 | 0.79 |
ENST00000359015.4 |
MAP3K5 |
mitogen-activated protein kinase kinase kinase 5 |
| chr12_-_50616122 | 0.78 |
ENST00000552823.1 ENST00000552909.1 |
LIMA1 |
LIM domain and actin binding 1 |
| chr11_+_111782934 | 0.77 |
ENST00000304298.3 |
HSPB2 |
Homo sapiens heat shock 27kDa protein 2 (HSPB2), mRNA. |
| chr3_+_35722487 | 0.77 |
ENST00000441454.1 |
ARPP21 |
cAMP-regulated phosphoprotein, 21kDa |
| chrX_-_107019181 | 0.76 |
ENST00000315660.4 ENST00000372384.2 ENST00000502650.1 ENST00000506724.1 |
TSC22D3 |
TSC22 domain family, member 3 |
| chrX_-_109561294 | 0.76 |
ENST00000372059.2 ENST00000262844.5 |
AMMECR1 |
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
| chr11_+_46299199 | 0.75 |
ENST00000529193.1 ENST00000288400.3 |
CREB3L1 |
cAMP responsive element binding protein 3-like 1 |
| chr12_+_79258547 | 0.75 |
ENST00000457153.2 |
SYT1 |
synaptotagmin I |
| chr8_-_59572093 | 0.74 |
ENST00000427130.2 |
NSMAF |
neutral sphingomyelinase (N-SMase) activation associated factor |
| chr11_-_119252359 | 0.74 |
ENST00000455332.2 |
USP2 |
ubiquitin specific peptidase 2 |
| chr19_+_14492247 | 0.74 |
ENST00000357355.3 ENST00000592261.2 ENST00000242786.5 |
CD97 |
CD97 molecule |
| chrX_-_137793826 | 0.74 |
ENST00000315930.6 |
FGF13 |
fibroblast growth factor 13 |
| chr1_-_161993422 | 0.72 |
ENST00000367940.2 |
OLFML2B |
olfactomedin-like 2B |
| chr8_-_101962777 | 0.72 |
ENST00000395951.3 |
YWHAZ |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr5_-_178772424 | 0.71 |
ENST00000251582.7 ENST00000274609.5 |
ADAMTS2 |
ADAM metallopeptidase with thrombospondin type 1 motif, 2 |
| chr11_-_64013288 | 0.70 |
ENST00000542235.1 |
PPP1R14B |
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr5_+_49962495 | 0.70 |
ENST00000515175.1 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
| chr22_+_19705928 | 0.69 |
ENST00000383045.3 ENST00000438754.2 |
SEPT5 |
septin 5 |
| chr4_-_186696425 | 0.69 |
ENST00000430503.1 ENST00000319454.6 ENST00000450341.1 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr7_-_27224795 | 0.69 |
ENST00000006015.3 |
HOXA11 |
homeobox A11 |
| chr12_-_77272765 | 0.69 |
ENST00000547435.1 ENST00000552330.1 ENST00000546966.1 ENST00000311083.5 |
CSRP2 |
cysteine and glycine-rich protein 2 |
| chr11_+_67776012 | 0.69 |
ENST00000539229.1 |
ALDH3B1 |
aldehyde dehydrogenase 3 family, member B1 |
| chr7_-_27224842 | 0.68 |
ENST00000517402.1 |
HOXA11 |
homeobox A11 |
| chr12_+_54378923 | 0.68 |
ENST00000303460.4 |
HOXC10 |
homeobox C10 |
| chr15_-_42749711 | 0.68 |
ENST00000565611.1 ENST00000263805.4 ENST00000565948.1 |
ZNF106 |
zinc finger protein 106 |
| chr20_-_44455976 | 0.68 |
ENST00000372555.3 |
TNNC2 |
troponin C type 2 (fast) |
| chr12_-_50616382 | 0.67 |
ENST00000552783.1 |
LIMA1 |
LIM domain and actin binding 1 |
| chr11_+_71938925 | 0.67 |
ENST00000538751.1 |
INPPL1 |
inositol polyphosphate phosphatase-like 1 |
| chr7_+_130794846 | 0.67 |
ENST00000421797.2 |
MKLN1 |
muskelin 1, intracellular mediator containing kelch motifs |
| chr11_-_2170786 | 0.66 |
ENST00000300632.5 |
IGF2 |
insulin-like growth factor 2 (somatomedin A) |
| chr1_-_144932316 | 0.65 |
ENST00000313431.9 |
PDE4DIP |
phosphodiesterase 4D interacting protein |
| chr16_-_89043605 | 0.65 |
ENST00000268679.4 |
CBFA2T3 |
core-binding factor, runt domain, alpha subunit 2; translocated to, 3 |
| chr17_+_60704762 | 0.65 |
ENST00000303375.5 |
MRC2 |
mannose receptor, C type 2 |
| chr1_-_144932014 | 0.64 |
ENST00000529945.1 |
PDE4DIP |
phosphodiesterase 4D interacting protein |
| chr7_+_73442487 | 0.64 |
ENST00000380575.4 ENST00000380584.4 ENST00000458204.1 ENST00000357036.5 ENST00000417091.1 ENST00000429192.1 ENST00000442310.1 ENST00000380553.4 ENST00000380576.5 ENST00000428787.1 ENST00000320399.6 |
ELN |
elastin |
| chr3_+_49059038 | 0.64 |
ENST00000451378.2 |
NDUFAF3 |
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr1_-_17304771 | 0.64 |
ENST00000375534.3 |
MFAP2 |
microfibrillar-associated protein 2 |
| chr2_-_197041193 | 0.64 |
ENST00000409228.1 |
STK17B |
serine/threonine kinase 17b |
| chr3_-_141868293 | 0.63 |
ENST00000317104.7 ENST00000494358.1 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr16_-_31085514 | 0.63 |
ENST00000300849.4 |
ZNF668 |
zinc finger protein 668 |
| chr6_+_41021027 | 0.61 |
ENST00000244669.2 |
APOBEC2 |
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2 |
| chr11_-_33891362 | 0.61 |
ENST00000395833.3 |
LMO2 |
LIM domain only 2 (rhombotin-like 1) |
| chr19_-_45996465 | 0.61 |
ENST00000430715.2 |
RTN2 |
reticulon 2 |
| chr15_+_43809797 | 0.61 |
ENST00000399453.1 ENST00000300231.5 |
MAP1A |
microtubule-associated protein 1A |
| chr1_+_180165672 | 0.61 |
ENST00000443059.1 |
QSOX1 |
quiescin Q6 sulfhydryl oxidase 1 |
| chr11_-_842509 | 0.60 |
ENST00000322028.4 |
POLR2L |
polymerase (RNA) II (DNA directed) polypeptide L, 7.6kDa |
| chr2_+_33359646 | 0.60 |
ENST00000390003.4 ENST00000418533.2 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
| chr12_-_104443890 | 0.59 |
ENST00000547583.1 ENST00000360814.4 ENST00000546851.1 |
GLT8D2 |
glycosyltransferase 8 domain containing 2 |
| chr2_+_65283529 | 0.59 |
ENST00000546106.1 ENST00000537589.1 ENST00000260569.4 |
CEP68 |
centrosomal protein 68kDa |
| chr2_+_238395803 | 0.59 |
ENST00000264605.3 |
MLPH |
melanophilin |
| chr6_-_41909466 | 0.59 |
ENST00000414200.2 |
CCND3 |
cyclin D3 |
| chr2_+_33359687 | 0.59 |
ENST00000402934.1 ENST00000404525.1 ENST00000407925.1 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
| chr11_+_1940786 | 0.59 |
ENST00000278317.6 ENST00000381561.4 ENST00000381548.3 ENST00000360603.3 ENST00000381549.3 |
TNNT3 |
troponin T type 3 (skeletal, fast) |
| chr17_-_39538550 | 0.58 |
ENST00000394001.1 |
KRT34 |
keratin 34 |
| chrX_-_77395186 | 0.58 |
ENST00000341864.5 |
TAF9B |
TAF9B RNA polymerase II, TATA box binding protein (TBP)-associated factor, 31kDa |
| chr12_+_122150646 | 0.58 |
ENST00000449592.2 |
TMEM120B |
transmembrane protein 120B |
| chr1_-_17445930 | 0.58 |
ENST00000375486.4 ENST00000375481.1 ENST00000444885.2 |
PADI2 |
peptidyl arginine deiminase, type II |
| chr11_-_67120974 | 0.57 |
ENST00000539074.1 ENST00000312419.3 |
POLD4 |
polymerase (DNA-directed), delta 4, accessory subunit |
| chr2_+_238395879 | 0.57 |
ENST00000445024.2 ENST00000338530.4 ENST00000409373.1 |
MLPH |
melanophilin |
| chr11_-_119252425 | 0.57 |
ENST00000260187.2 |
USP2 |
ubiquitin specific peptidase 2 |
| chr4_-_186697044 | 0.57 |
ENST00000437304.2 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr1_+_165600083 | 0.57 |
ENST00000367889.3 |
MGST3 |
microsomal glutathione S-transferase 3 |
| chr13_-_24007815 | 0.56 |
ENST00000382298.3 |
SACS |
spastic ataxia of Charlevoix-Saguenay (sacsin) |
| chr12_-_50677255 | 0.56 |
ENST00000551691.1 ENST00000394943.3 ENST00000341247.4 |
LIMA1 |
LIM domain and actin binding 1 |
| chr21_+_27011584 | 0.56 |
ENST00000400532.1 ENST00000480456.1 ENST00000312957.5 |
JAM2 |
junctional adhesion molecule 2 |
| chr9_-_16727978 | 0.56 |
ENST00000418777.1 ENST00000468187.2 |
BNC2 |
basonuclin 2 |
| chr11_-_122930121 | 0.55 |
ENST00000524552.1 |
HSPA8 |
heat shock 70kDa protein 8 |
| chr10_-_62704005 | 0.55 |
ENST00000337910.5 |
RHOBTB1 |
Rho-related BTB domain containing 1 |
| chr3_+_183894566 | 0.55 |
ENST00000439647.1 |
AP2M1 |
adaptor-related protein complex 2, mu 1 subunit |
| chr10_-_79398127 | 0.54 |
ENST00000372443.1 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr1_+_165600436 | 0.54 |
ENST00000367888.4 ENST00000367885.1 ENST00000367884.2 |
MGST3 |
microsomal glutathione S-transferase 3 |
Gene Ontology Analysis
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.9 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 3.0 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 0.7 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 5.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 1.7 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 5.0 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 0.7 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 0.3 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 0.4 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.1 | 3.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 1.2 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 1.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 6.0 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 3.3 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.1 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.9 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.4 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.2 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 1.1 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 1.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 1.2 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 2.6 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 1.6 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 2.8 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.9 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.9 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.4 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 1.0 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.5 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 4.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.3 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.6 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.2 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.5 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.1 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 1.0 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.3 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.3 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 1.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.3 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.6 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.1 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 12.6 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 1.1 | 3.3 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.5 | 1.6 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.5 | 2.6 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.5 | 5.6 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.4 | 1.2 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.3 | 6.3 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.3 | 2.3 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.3 | 1.6 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.3 | 3.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.3 | 1.4 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.3 | 2.4 | GO:0043426 | MRF binding(GO:0043426) |
| 0.3 | 1.0 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 0.3 | 2.0 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.2 | 1.9 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 1.7 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.2 | 0.9 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.2 | 1.4 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.2 | 1.6 | GO:0071253 | connexin binding(GO:0071253) |
| 0.2 | 0.9 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 21.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.2 | 2.6 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.2 | 0.6 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.2 | 0.6 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.2 | 1.0 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.2 | 0.9 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.2 | 3.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.2 | 1.3 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.2 | 2.9 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.2 | 2.9 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.2 | 0.5 | GO:0004515 | nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.2 | 1.0 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.2 | 0.5 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.2 | 0.6 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) thiol oxidase activity(GO:0016972) |
| 0.1 | 1.4 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.1 | 1.0 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.1 | 4.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.4 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.1 | 0.5 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.1 | 0.6 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.1 | 1.4 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.5 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 0.5 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.1 | 0.4 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 0.3 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 0.8 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 0.3 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.1 | 1.2 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.1 | 0.3 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.1 | 0.5 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.3 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.1 | 0.4 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.1 | 1.8 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 1.0 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.1 | 0.3 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.1 | 0.4 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 0.4 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.1 | 0.9 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 0.3 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.1 | 0.3 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.1 | 0.7 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 1.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 0.4 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.4 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 2.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.3 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.1 | 0.1 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 1.1 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.1 | 0.6 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.1 | 1.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 0.3 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.1 | 0.4 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.1 | 0.2 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.2 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 2.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 0.2 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.1 | 1.5 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 0.2 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.1 | 2.2 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 0.2 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 1.0 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.1 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.2 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 2.8 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 1.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.5 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.8 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.5 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 1.0 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 1.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 2.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.4 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 5.5 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.3 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 1.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 2.3 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.3 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 6.2 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.2 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.0 | 0.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 2.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.7 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.6 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.4 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.4 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 1.5 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 2.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.1 | GO:0070643 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.3 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.0 | 0.4 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.6 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 0.2 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.1 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.0 | 0.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.7 | GO:0043394 | proteoglycan binding(GO:0043394) |
| 0.0 | 0.2 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.5 | GO:0090079 | translation repressor activity, nucleic acid binding(GO:0000900) translation regulator activity, nucleic acid binding(GO:0090079) |
| 0.0 | 0.7 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 1.2 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.3 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.2 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.3 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.2 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.4 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.2 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.9 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.3 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.7 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 0.2 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.1 | GO:1901474 | thiamine transmembrane transporter activity(GO:0015234) thiamine uptake transmembrane transporter activity(GO:0015403) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 1.5 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 1.2 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 1.0 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.7 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.3 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.3 | GO:0008603 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.1 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 2.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.3 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 2.2 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.1 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.7 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.1 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.5 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.9 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 1.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.3 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.2 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.4 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.8 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 0.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.4 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 0.1 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.1 | GO:0017091 | AU-rich element binding(GO:0017091) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 6.0 | GO:1900075 | regulation of neuromuscular synaptic transmission(GO:1900073) positive regulation of neuromuscular synaptic transmission(GO:1900075) |
| 1.1 | 3.3 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 1.0 | 4.0 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 1.0 | 3.0 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.9 | 3.4 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.8 | 1.6 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.5 | 1.6 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.5 | 1.5 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.5 | 1.4 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.5 | 1.4 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.4 | 32.6 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.4 | 5.9 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 0.4 | 1.7 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.4 | 2.8 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.4 | 2.0 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.4 | 1.6 | GO:0014819 | regulation of skeletal muscle contraction(GO:0014819) |
| 0.4 | 2.6 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.4 | 1.8 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.3 | 0.9 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.3 | 2.4 | GO:0070235 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.3 | 1.2 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 0.3 | 2.8 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.2 | 1.0 | GO:0044111 | development involved in symbiotic interaction(GO:0044111) |
| 0.2 | 3.1 | GO:0035826 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.2 | 1.0 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.2 | 0.7 | GO:1901899 | positive regulation of relaxation of cardiac muscle(GO:1901899) regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 0.2 | 3.4 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.2 | 1.7 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.2 | 0.6 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.2 | 1.0 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.2 | 2.2 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.2 | 0.8 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.2 | 0.6 | GO:1901624 | negative regulation of lymphocyte chemotaxis(GO:1901624) |
| 0.2 | 0.8 | GO:2001153 | regulation of renal water transport(GO:2001151) positive regulation of renal water transport(GO:2001153) |
| 0.2 | 3.5 | GO:0045199 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.2 | 1.6 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.2 | 0.7 | GO:1904616 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.2 | 1.9 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.2 | 1.6 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.2 | 0.3 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
| 0.2 | 1.2 | GO:0071694 | sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) |
| 0.2 | 0.8 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.2 | 2.6 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.2 | 0.6 | GO:0006051 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.2 | 1.0 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.2 | 0.5 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 | 3.1 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 0.7 | GO:1990834 | response to odorant(GO:1990834) |
| 0.1 | 0.4 | GO:1905154 | negative regulation of membrane invagination(GO:1905154) |
| 0.1 | 0.7 | GO:0015862 | uridine transport(GO:0015862) |
| 0.1 | 0.4 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.1 | 0.4 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.1 | 2.0 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 0.5 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 0.1 | 1.5 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 0.4 | GO:1904772 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.1 | 0.9 | GO:1902302 | regulation of potassium ion export(GO:1902302) |
| 0.1 | 0.5 | GO:0036496 | regulation of translational initiation by eIF2 alpha dephosphorylation(GO:0036496) |
| 0.1 | 0.4 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.1 | 0.2 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.1 | 0.4 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.1 | 1.1 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 0.5 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.1 | 0.7 | GO:0052551 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 0.1 | 0.8 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.1 | 0.6 | GO:0061738 | late endosomal microautophagy(GO:0061738) negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.1 | 0.9 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.1 | 0.4 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.1 | 1.6 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.1 | 0.3 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.1 | 0.5 | GO:0038170 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.1 | 0.5 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.1 | 0.6 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.1 | 2.3 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.1 | 0.9 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 | 0.3 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.1 | 0.9 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.1 | 0.6 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.1 | 0.8 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.1 | 0.4 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.3 | GO:0006260 | DNA replication(GO:0006260) |
| 0.1 | 0.4 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 0.3 | GO:0021558 | trochlear nerve development(GO:0021558) |
| 0.1 | 2.6 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.1 | 1.3 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 0.3 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.1 | 0.4 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.1 | 1.6 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.3 | GO:0035854 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) |
| 0.1 | 0.2 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.1 | 0.4 | GO:2001046 | positive regulation of integrin-mediated signaling pathway(GO:2001046) |
| 0.1 | 0.9 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.4 | GO:0097688 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.1 | 0.9 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.1 | 0.4 | GO:1905040 | vestibulocochlear nerve structural organization(GO:0021649) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) endothelial tip cell fate specification(GO:0097102) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.1 | 0.8 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.1 | 0.3 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.1 | 0.3 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.1 | 0.2 | GO:0032252 | secretory granule localization(GO:0032252) |
| 0.1 | 0.3 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.1 | 1.0 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.1 | 0.9 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.1 | 1.2 | GO:0007501 | mesodermal cell fate specification(GO:0007501) |
| 0.1 | 2.6 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 0.3 | GO:0010193 | response to ozone(GO:0010193) |
| 0.1 | 0.7 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.1 | 0.1 | GO:0002331 | pre-B cell differentiation(GO:0002329) pre-B cell allelic exclusion(GO:0002331) |
| 0.1 | 1.2 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.1 | 0.1 | GO:0061461 | lysine import(GO:0034226) L-lysine import(GO:0061461) L-lysine import into cell(GO:1903410) |
| 0.1 | 0.4 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 0.7 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.1 | 0.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 1.4 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 0.5 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.4 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.1 | 0.5 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 0.5 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.1 | 0.9 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.1 | 0.1 | GO:0032804 | negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) |
| 0.1 | 0.6 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.1 | 0.8 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 0.9 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 0.7 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.1 | 0.1 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.1 | 0.4 | GO:0030421 | defecation(GO:0030421) |
| 0.1 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.1 | 0.4 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.1 | 0.2 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 | 0.2 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.1 | 1.4 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.1 | 0.7 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.2 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.1 | 0.3 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.1 | 0.3 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.1 | 0.2 | GO:0097068 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.1 | 0.1 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.1 | 0.4 | GO:0002326 | B cell lineage commitment(GO:0002326) immunoglobulin V(D)J recombination(GO:0033152) |
| 0.1 | 0.1 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.1 | 0.6 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.1 | 0.2 | GO:0060437 | lung growth(GO:0060437) |
| 0.1 | 0.3 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.1 | 0.2 | GO:0070213 | negative regulation of sister chromatid cohesion(GO:0045875) protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 | 0.3 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.1 | 0.4 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 0.2 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.0 | 0.2 | GO:1901098 | regulation of autophagosome maturation(GO:1901096) positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.2 | GO:0006238 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.0 | 0.7 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.2 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.5 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.2 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 1.4 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 1.4 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.2 | GO:1903436 | regulation of cytokinetic process(GO:0032954) positive regulation of centriole replication(GO:0046601) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.0 | 1.7 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.1 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.0 | 0.6 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.0 | 1.8 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 3.4 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.2 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.0 | GO:0072053 | renal inner medulla development(GO:0072053) renal outer medulla development(GO:0072054) |
| 0.0 | 0.0 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.1 | GO:1903925 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.0 | 0.0 | GO:0048563 | post-embryonic organ morphogenesis(GO:0048563) |
| 0.0 | 1.5 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.0 | 0.3 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.0 | 0.3 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.3 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.0 | 0.6 | GO:0048853 | forebrain morphogenesis(GO:0048853) |
| 0.0 | 1.8 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.0 | 0.4 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 1.5 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.0 | 0.6 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 2.6 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 1.0 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.6 | GO:0043586 | tongue development(GO:0043586) |
| 0.0 | 0.2 | GO:1990262 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 0.0 | 0.2 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.3 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.0 | 0.1 | GO:1904637 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.0 | 0.5 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 0.0 | 0.5 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.1 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 | 0.2 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.1 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.8 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 0.2 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.2 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.3 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.1 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.1 | GO:1904261 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.0 | 0.3 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.1 | GO:0036378 | calcitriol biosynthetic process from calciol(GO:0036378) vitamin D3 metabolic process(GO:0070640) |
| 0.0 | 0.2 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.3 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.3 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0045820 | negative regulation of glycolytic process(GO:0045820) negative regulation of cofactor metabolic process(GO:0051195) negative regulation of coenzyme metabolic process(GO:0051198) |
| 0.0 | 0.5 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 4.4 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 0.3 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 1.0 | GO:0003012 | muscle system process(GO:0003012) |
| 0.0 | 1.5 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.1 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.6 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.1 | GO:1902723 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.0 | 0.2 | GO:0034770 | histone H4-K20 methylation(GO:0034770) |
| 0.0 | 0.1 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.0 | 0.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.1 | GO:0061484 | inner cell mass cell differentiation(GO:0001826) hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.1 | GO:0015670 | carbon dioxide transport(GO:0015670) saliva secretion(GO:0046541) |
| 0.0 | 0.5 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 1.5 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.0 | 0.1 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 0.7 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.3 | GO:1903362 | regulation of cellular protein catabolic process(GO:1903362) |
| 0.0 | 0.4 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.0 | 0.4 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.5 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.4 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.3 | GO:0034134 | toll-like receptor 2 signaling pathway(GO:0034134) |
| 0.0 | 0.2 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.2 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.4 | GO:0021511 | spinal cord patterning(GO:0021511) |
| 0.0 | 0.4 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.3 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.1 | GO:0061087 | negative regulation of histone H3-K27 methylation(GO:0061086) positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 | 0.1 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.0 | 0.1 | GO:0001806 | type IV hypersensitivity(GO:0001806) regulation of type IV hypersensitivity(GO:0001807) |
| 0.0 | 0.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.4 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.0 | 0.3 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.0 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.1 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.0 | 0.8 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.2 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.6 | GO:0006266 | DNA ligation(GO:0006266) |
| 0.0 | 0.1 | GO:0071934 | thiamine transport(GO:0015888) thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.2 | GO:0051412 | response to corticosterone(GO:0051412) |
| 0.0 | 0.2 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.0 | 0.1 | GO:0043497 | regulation of protein heterodimerization activity(GO:0043497) |
| 0.0 | 0.1 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.0 | 0.2 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.1 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.7 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 2.5 | GO:0001764 | neuron migration(GO:0001764) |
| 0.0 | 0.3 | GO:0009299 | mRNA transcription(GO:0009299) |
| 0.0 | 2.2 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 | 0.4 | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.0 | 0.4 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.1 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.0 | 1.0 | GO:1902400 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) signal transduction involved in mitotic cell cycle checkpoint(GO:0072413) signal transduction involved in mitotic G1 DNA damage checkpoint(GO:0072431) intracellular signal transduction involved in G1 DNA damage checkpoint(GO:1902400) signal transduction involved in mitotic DNA damage checkpoint(GO:1902402) signal transduction involved in mitotic DNA integrity checkpoint(GO:1902403) |
| 0.0 | 0.1 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 1.3 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.4 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.3 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 | 0.3 | GO:0045765 | regulation of angiogenesis(GO:0045765) |
| 0.0 | 0.5 | GO:0048255 | mRNA stabilization(GO:0048255) |
| 0.0 | 0.1 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.0 | GO:0030222 | eosinophil differentiation(GO:0030222) |
| 0.0 | 0.1 | GO:0001824 | blastocyst development(GO:0001824) |
| 0.0 | 0.2 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.1 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.0 | 0.1 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.5 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.3 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.1 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.3 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
| 0.0 | 0.1 | GO:0016102 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.0 | 0.7 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.9 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.1 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.1 | GO:2000501 | regulation of natural killer cell chemotaxis(GO:2000501) positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.0 | GO:0070671 | interleukin-12-mediated signaling pathway(GO:0035722) response to interleukin-12(GO:0070671) cellular response to interleukin-12(GO:0071349) |
| 0.0 | 0.2 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.0 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.1 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 1.5 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.1 | GO:0043542 | endothelial cell migration(GO:0043542) |
| 0.0 | 0.0 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.5 | GO:0006221 | pyrimidine nucleotide biosynthetic process(GO:0006221) |
| 0.0 | 0.4 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.8 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.0 | 0.1 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.0 | 0.1 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.0 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.2 | GO:0006379 | mRNA cleavage(GO:0006379) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.7 | 2.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.7 | 4.0 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.5 | 6.4 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.4 | 2.6 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.4 | 7.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.4 | 4.6 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.4 | 7.0 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.4 | 7.6 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.3 | 1.6 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.3 | 1.4 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.3 | 3.1 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.3 | 16.3 | GO:0031672 | A band(GO:0031672) |
| 0.2 | 3.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.2 | 0.8 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.2 | 3.8 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.2 | 1.3 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 1.0 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 0.6 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 1.3 | GO:0031674 | I band(GO:0031674) |
| 0.1 | 0.9 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 19.3 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 0.6 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.1 | 3.4 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 2.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 1.0 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 0.3 | GO:0005761 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.1 | 1.3 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 2.8 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 0.4 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.8 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.1 | 0.6 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.7 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 0.5 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.1 | 0.2 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 0.1 | 1.5 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 0.8 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.7 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 0.2 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.1 | 2.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 0.2 | GO:0020003 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.1 | 0.5 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 1.5 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.4 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 0.3 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.3 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.1 | 0.3 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.3 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.9 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.7 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.2 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.1 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.1 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.2 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.5 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.6 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.4 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.6 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.3 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.8 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.3 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.0 | 1.0 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 1.8 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 1.2 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.8 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.5 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.4 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.0 | 0.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.5 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.3 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.5 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.6 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.3 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 1.8 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.6 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.4 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 2.3 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.5 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.0 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 2.5 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.2 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.6 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 1.3 | GO:0030016 | myofibril(GO:0030016) |
| 0.0 | 0.6 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 1.9 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 3.0 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 0.1 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.4 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.9 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.1 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.2 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 2.3 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.2 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.0 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.0 | 0.3 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 1.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.1 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 1.8 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.1 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 7.0 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.2 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.0 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.3 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 3.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.1 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 1.3 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.6 | GO:0043195 | terminal bouton(GO:0043195) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 35.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 4.6 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 0.9 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.1 | 1.5 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.1 | 1.9 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 2.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 0.9 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.7 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.1 | 1.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.7 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 1.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 3.9 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.1 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 2.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.1 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.7 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 1.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.7 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.6 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 3.4 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 2.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.3 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.8 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 1.0 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.6 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 1.0 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 3.5 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.8 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 1.3 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.9 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.4 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.5 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.6 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 1.0 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.2 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.3 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.6 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.1 | REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | Genes involved in p53-Independent G1/S DNA damage checkpoint |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.0 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.2 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.1 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.1 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.3 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 1.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |