Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
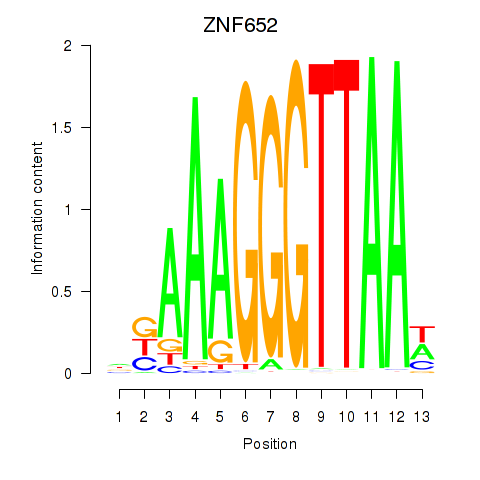
Results for ZNF652
Z-value: 2.37
Transcription factors associated with ZNF652
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF652
|
ENSG00000198740.4 | ZNF652 |
Activity profile of ZNF652 motif
Sorted Z-values of ZNF652 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF652
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_157154578 | 10.65 |
ENST00000295927.3 |
PTX3 |
pentraxin 3, long |
| chr1_-_156647189 | 9.52 |
ENST00000368223.3 |
NES |
nestin |
| chr2_-_190044480 | 7.76 |
ENST00000374866.3 |
COL5A2 |
collagen, type V, alpha 2 |
| chr16_+_31483451 | 6.22 |
ENST00000565360.1 ENST00000361773.3 |
TGFB1I1 |
transforming growth factor beta 1 induced transcript 1 |
| chr19_+_41725088 | 5.97 |
ENST00000301178.4 |
AXL |
AXL receptor tyrosine kinase |
| chr11_-_89224508 | 5.56 |
ENST00000525196.1 |
NOX4 |
NADPH oxidase 4 |
| chr16_+_31483374 | 5.44 |
ENST00000394863.3 |
TGFB1I1 |
transforming growth factor beta 1 induced transcript 1 |
| chr8_-_23261589 | 5.29 |
ENST00000524168.1 ENST00000523833.2 ENST00000519243.1 ENST00000389131.3 |
LOXL2 |
lysyl oxidase-like 2 |
| chr5_-_54281407 | 5.20 |
ENST00000381403.4 |
ESM1 |
endothelial cell-specific molecule 1 |
| chr11_-_89224638 | 5.18 |
ENST00000535633.1 ENST00000263317.4 |
NOX4 |
NADPH oxidase 4 |
| chr4_-_99578789 | 5.04 |
ENST00000511651.1 ENST00000505184.1 |
TSPAN5 |
tetraspanin 5 |
| chr11_-_89224488 | 4.82 |
ENST00000534731.1 ENST00000527626.1 |
NOX4 |
NADPH oxidase 4 |
| chr3_+_158991025 | 4.80 |
ENST00000337808.6 |
IQCJ-SCHIP1 |
IQCJ-SCHIP1 readthrough |
| chr11_-_35547151 | 4.71 |
ENST00000378878.3 ENST00000529303.1 ENST00000278360.3 |
PAMR1 |
peptidase domain containing associated with muscle regeneration 1 |
| chr13_-_38172863 | 4.62 |
ENST00000541481.1 ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN |
periostin, osteoblast specific factor |
| chr11_-_89224299 | 4.58 |
ENST00000343727.5 ENST00000531342.1 ENST00000375979.3 |
NOX4 |
NADPH oxidase 4 |
| chr11_-_35547572 | 4.58 |
ENST00000378880.2 |
PAMR1 |
peptidase domain containing associated with muscle regeneration 1 |
| chr11_-_89224139 | 4.52 |
ENST00000413594.2 |
NOX4 |
NADPH oxidase 4 |
| chr2_-_161350305 | 4.39 |
ENST00000348849.3 |
RBMS1 |
RNA binding motif, single stranded interacting protein 1 |
| chr4_-_99578776 | 4.30 |
ENST00000515287.1 |
TSPAN5 |
tetraspanin 5 |
| chr14_+_85996471 | 4.10 |
ENST00000330753.4 |
FLRT2 |
fibronectin leucine rich transmembrane protein 2 |
| chr2_-_188378368 | 4.07 |
ENST00000392365.1 ENST00000435414.1 |
TFPI |
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr5_+_135394840 | 3.93 |
ENST00000503087.1 |
TGFBI |
transforming growth factor, beta-induced, 68kDa |
| chr11_-_119293872 | 3.37 |
ENST00000524970.1 |
THY1 |
Thy-1 cell surface antigen |
| chr11_-_89223883 | 3.36 |
ENST00000528341.1 |
NOX4 |
NADPH oxidase 4 |
| chr1_+_78470530 | 3.34 |
ENST00000370763.5 |
DNAJB4 |
DnaJ (Hsp40) homolog, subfamily B, member 4 |
| chr11_-_111782696 | 3.34 |
ENST00000227251.3 ENST00000526180.1 |
CRYAB |
crystallin, alpha B |
| chr1_-_193155729 | 3.28 |
ENST00000367434.4 |
B3GALT2 |
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chr11_+_19798964 | 3.28 |
ENST00000527559.2 |
NAV2 |
neuron navigator 2 |
| chr2_-_218808771 | 3.05 |
ENST00000449814.1 ENST00000171887.4 |
TNS1 |
tensin 1 |
| chr20_-_36793774 | 3.05 |
ENST00000361475.2 |
TGM2 |
transglutaminase 2 |
| chr5_-_16936340 | 2.89 |
ENST00000507288.1 ENST00000513610.1 |
MYO10 |
myosin X |
| chr11_-_111782484 | 2.83 |
ENST00000533971.1 |
CRYAB |
crystallin, alpha B |
| chr1_-_150780757 | 2.81 |
ENST00000271651.3 |
CTSK |
cathepsin K |
| chr19_-_44174330 | 2.77 |
ENST00000340093.3 |
PLAUR |
plasminogen activator, urokinase receptor |
| chr4_-_143226979 | 2.71 |
ENST00000514525.1 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr10_-_15413035 | 2.71 |
ENST00000378116.4 ENST00000455654.1 |
FAM171A1 |
family with sequence similarity 171, member A1 |
| chr14_-_105420241 | 2.70 |
ENST00000557457.1 |
AHNAK2 |
AHNAK nucleoprotein 2 |
| chr16_+_3068393 | 2.63 |
ENST00000573001.1 |
TNFRSF12A |
tumor necrosis factor receptor superfamily, member 12A |
| chr19_-_44174305 | 2.58 |
ENST00000601723.1 ENST00000339082.3 |
PLAUR |
plasminogen activator, urokinase receptor |
| chr14_+_85996507 | 2.56 |
ENST00000554746.1 |
FLRT2 |
fibronectin leucine rich transmembrane protein 2 |
| chr1_+_101185290 | 2.41 |
ENST00000370119.4 ENST00000347652.2 ENST00000294728.2 ENST00000370115.1 |
VCAM1 |
vascular cell adhesion molecule 1 |
| chr1_-_33168336 | 2.38 |
ENST00000373484.3 |
SYNC |
syncoilin, intermediate filament protein |
| chr19_+_41725140 | 2.36 |
ENST00000359092.3 |
AXL |
AXL receptor tyrosine kinase |
| chr3_-_127441406 | 2.34 |
ENST00000487473.1 ENST00000484451.1 |
MGLL |
monoglyceride lipase |
| chr22_+_38609538 | 2.30 |
ENST00000407965.1 |
MAFF |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
| chr5_+_125758813 | 2.30 |
ENST00000285689.3 ENST00000515200.1 |
GRAMD3 |
GRAM domain containing 3 |
| chr11_-_63381925 | 2.29 |
ENST00000415826.1 |
PLA2G16 |
phospholipase A2, group XVI |
| chr11_-_86383157 | 2.28 |
ENST00000393324.3 |
ME3 |
malic enzyme 3, NADP(+)-dependent, mitochondrial |
| chr16_-_79634595 | 2.21 |
ENST00000326043.4 ENST00000393350.1 |
MAF |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog |
| chr5_+_125758865 | 2.21 |
ENST00000542322.1 ENST00000544396.1 |
GRAMD3 |
GRAM domain containing 3 |
| chr9_-_117853297 | 2.20 |
ENST00000542877.1 ENST00000537320.1 ENST00000341037.4 |
TNC |
tenascin C |
| chr14_-_88459503 | 2.13 |
ENST00000393568.4 ENST00000261304.2 |
GALC |
galactosylceramidase |
| chr14_+_59104741 | 2.13 |
ENST00000395153.3 ENST00000335867.4 |
DACT1 |
dishevelled-binding antagonist of beta-catenin 1 |
| chr12_-_6451186 | 2.11 |
ENST00000540022.1 ENST00000536194.1 |
TNFRSF1A |
tumor necrosis factor receptor superfamily, member 1A |
| chr10_+_11060004 | 2.09 |
ENST00000542579.1 ENST00000399850.3 ENST00000417956.2 |
CELF2 |
CUGBP, Elav-like family member 2 |
| chr4_-_77134742 | 2.06 |
ENST00000452464.2 |
SCARB2 |
scavenger receptor class B, member 2 |
| chr11_-_86383370 | 2.05 |
ENST00000526834.1 ENST00000359636.2 |
ME3 |
malic enzyme 3, NADP(+)-dependent, mitochondrial |
| chr11_+_111782934 | 2.01 |
ENST00000304298.3 |
HSPB2 |
Homo sapiens heat shock 27kDa protein 2 (HSPB2), mRNA. |
| chr15_+_63569785 | 1.97 |
ENST00000380343.4 ENST00000560353.1 |
APH1B |
APH1B gamma secretase subunit |
| chr12_-_123756781 | 1.86 |
ENST00000544658.1 |
CDK2AP1 |
cyclin-dependent kinase 2 associated protein 1 |
| chr9_-_35111570 | 1.86 |
ENST00000378561.1 ENST00000603301.1 |
FAM214B |
family with sequence similarity 214, member B |
| chr20_-_36793663 | 1.81 |
ENST00000536701.1 ENST00000536724.1 |
TGM2 |
transglutaminase 2 |
| chr12_-_123756687 | 1.75 |
ENST00000261692.2 |
CDK2AP1 |
cyclin-dependent kinase 2 associated protein 1 |
| chr15_+_68871569 | 1.74 |
ENST00000566799.1 |
CORO2B |
coronin, actin binding protein, 2B |
| chr9_-_35111420 | 1.62 |
ENST00000378557.1 |
FAM214B |
family with sequence similarity 214, member B |
| chr1_+_145293371 | 1.59 |
ENST00000342960.5 |
NBPF10 |
neuroblastoma breakpoint family, member 10 |
| chr12_+_56546223 | 1.58 |
ENST00000550443.1 ENST00000207437.5 |
MYL6B |
myosin, light chain 6B, alkali, smooth muscle and non-muscle |
| chr6_-_52441713 | 1.58 |
ENST00000182527.3 |
TRAM2 |
translocation associated membrane protein 2 |
| chr7_-_30066233 | 1.57 |
ENST00000222803.5 |
FKBP14 |
FK506 binding protein 14, 22 kDa |
| chr14_+_24867992 | 1.56 |
ENST00000382554.3 |
NYNRIN |
NYN domain and retroviral integrase containing |
| chr8_-_131455835 | 1.51 |
ENST00000518721.1 |
ASAP1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
| chr8_-_17555164 | 1.49 |
ENST00000297488.6 |
MTUS1 |
microtubule associated tumor suppressor 1 |
| chr10_+_11059826 | 1.49 |
ENST00000450189.1 |
CELF2 |
CUGBP, Elav-like family member 2 |
| chr12_-_6451235 | 1.46 |
ENST00000440083.2 ENST00000162749.2 |
TNFRSF1A |
tumor necrosis factor receptor superfamily, member 1A |
| chr6_+_39760783 | 1.42 |
ENST00000398904.2 ENST00000538976.1 |
DAAM2 |
dishevelled associated activator of morphogenesis 2 |
| chr15_+_25200074 | 1.39 |
ENST00000390687.4 ENST00000584968.1 ENST00000346403.6 ENST00000554227.2 |
SNRPN |
small nuclear ribonucleoprotein polypeptide N |
| chr12_+_56546363 | 1.39 |
ENST00000551834.1 ENST00000552568.1 |
MYL6B |
myosin, light chain 6B, alkali, smooth muscle and non-muscle |
| chr6_-_131321863 | 1.37 |
ENST00000528282.1 |
EPB41L2 |
erythrocyte membrane protein band 4.1-like 2 |
| chr5_-_157002775 | 1.34 |
ENST00000257527.4 |
ADAM19 |
ADAM metallopeptidase domain 19 |
| chr11_-_104840093 | 1.30 |
ENST00000417440.2 ENST00000444739.2 |
CASP4 |
caspase 4, apoptosis-related cysteine peptidase |
| chr16_-_66584059 | 1.26 |
ENST00000417693.3 ENST00000544898.1 ENST00000569718.1 ENST00000527284.1 ENST00000299697.7 ENST00000451102.2 |
TK2 |
thymidine kinase 2, mitochondrial |
| chr4_-_186732048 | 1.23 |
ENST00000448662.2 ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr22_-_37880543 | 1.22 |
ENST00000442496.1 |
MFNG |
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr8_-_101962777 | 1.21 |
ENST00000395951.3 |
YWHAZ |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr1_-_208417620 | 1.15 |
ENST00000367033.3 |
PLXNA2 |
plexin A2 |
| chr7_+_23286182 | 1.15 |
ENST00000258733.4 ENST00000381990.2 ENST00000409458.3 ENST00000539136.1 ENST00000453162.2 |
GPNMB |
glycoprotein (transmembrane) nmb |
| chr4_-_90756769 | 1.13 |
ENST00000345009.4 ENST00000505199.1 ENST00000502987.1 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
| chr1_+_82266053 | 1.08 |
ENST00000370715.1 ENST00000370713.1 ENST00000319517.6 ENST00000370717.2 ENST00000394879.1 ENST00000271029.4 ENST00000335786.5 |
LPHN2 |
latrophilin 2 |
| chr7_+_12726474 | 1.08 |
ENST00000396662.1 ENST00000356797.3 ENST00000396664.2 |
ARL4A |
ADP-ribosylation factor-like 4A |
| chr1_+_145293114 | 1.08 |
ENST00000369338.1 |
NBPF10 |
neuroblastoma breakpoint family, member 10 |
| chr22_+_29702572 | 1.07 |
ENST00000407647.2 ENST00000416823.1 ENST00000428622.1 |
GAS2L1 |
growth arrest-specific 2 like 1 |
| chr4_-_143227088 | 1.07 |
ENST00000511838.1 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr7_+_12727250 | 1.06 |
ENST00000404894.1 |
ARL4A |
ADP-ribosylation factor-like 4A |
| chr19_+_17326191 | 1.04 |
ENST00000595101.1 ENST00000596136.1 ENST00000379776.4 |
USE1 |
unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
| chr4_-_90757364 | 1.02 |
ENST00000508895.1 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
| chr16_-_49890016 | 1.00 |
ENST00000563137.2 |
ZNF423 |
zinc finger protein 423 |
| chr15_+_71185148 | 0.98 |
ENST00000443425.2 ENST00000560755.1 |
LRRC49 |
leucine rich repeat containing 49 |
| chr1_+_169077172 | 0.97 |
ENST00000499679.3 |
ATP1B1 |
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr17_-_19290117 | 0.94 |
ENST00000497081.2 |
MFAP4 |
microfibrillar-associated protein 4 |
| chr10_+_124320195 | 0.90 |
ENST00000359586.6 |
DMBT1 |
deleted in malignant brain tumors 1 |
| chr3_+_113616317 | 0.90 |
ENST00000440446.2 ENST00000488680.1 |
GRAMD1C |
GRAM domain containing 1C |
| chr20_-_60942361 | 0.90 |
ENST00000252999.3 |
LAMA5 |
laminin, alpha 5 |
| chr4_+_129730947 | 0.83 |
ENST00000452328.2 ENST00000504089.1 |
PHF17 |
jade family PHD finger 1 |
| chr7_+_80231466 | 0.80 |
ENST00000309881.7 ENST00000534394.1 |
CD36 |
CD36 molecule (thrombospondin receptor) |
| chr15_-_63449663 | 0.79 |
ENST00000439025.1 |
RPS27L |
ribosomal protein S27-like |
| chr15_+_68871308 | 0.78 |
ENST00000261861.5 |
CORO2B |
coronin, actin binding protein, 2B |
| chr9_-_2844058 | 0.77 |
ENST00000397885.2 |
KIAA0020 |
KIAA0020 |
| chr2_+_138722028 | 0.77 |
ENST00000280096.5 |
HNMT |
histamine N-methyltransferase |
| chr2_-_86564776 | 0.75 |
ENST00000165698.5 ENST00000541910.1 ENST00000535845.1 |
REEP1 |
receptor accessory protein 1 |
| chr16_+_67927147 | 0.71 |
ENST00000291041.5 |
PSKH1 |
protein serine kinase H1 |
| chr2_+_162016916 | 0.71 |
ENST00000405852.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr10_+_51187938 | 0.68 |
ENST00000311663.5 |
FAM21D |
family with sequence similarity 21, member D |
| chr8_-_101734308 | 0.68 |
ENST00000519004.1 ENST00000519363.1 ENST00000520142.1 |
PABPC1 |
poly(A) binding protein, cytoplasmic 1 |
| chr19_-_10530784 | 0.67 |
ENST00000593124.1 |
CDC37 |
cell division cycle 37 |
| chr14_+_79745746 | 0.67 |
ENST00000281127.7 |
NRXN3 |
neurexin 3 |
| chr2_+_152214098 | 0.65 |
ENST00000243347.3 |
TNFAIP6 |
tumor necrosis factor, alpha-induced protein 6 |
| chr2_+_177053307 | 0.63 |
ENST00000331462.4 |
HOXD1 |
homeobox D1 |
| chrX_-_102943022 | 0.62 |
ENST00000433176.2 |
MORF4L2 |
mortality factor 4 like 2 |
| chr11_-_130298888 | 0.62 |
ENST00000257359.6 |
ADAMTS8 |
ADAM metallopeptidase with thrombospondin type 1 motif, 8 |
| chr4_-_74486217 | 0.62 |
ENST00000335049.5 ENST00000307439.5 |
RASSF6 |
Ras association (RalGDS/AF-6) domain family member 6 |
| chrX_-_102942961 | 0.61 |
ENST00000434230.1 ENST00000418819.1 ENST00000360458.1 |
MORF4L2 |
mortality factor 4 like 2 |
| chr3_+_46449049 | 0.59 |
ENST00000357392.4 ENST00000400880.3 ENST00000433848.1 |
CCRL2 |
chemokine (C-C motif) receptor-like 2 |
| chr2_+_162016804 | 0.59 |
ENST00000392749.2 ENST00000440506.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr2_+_99225018 | 0.58 |
ENST00000357765.2 ENST00000409975.1 |
UNC50 |
unc-50 homolog (C. elegans) |
| chr17_-_42908155 | 0.57 |
ENST00000426548.1 ENST00000590758.1 ENST00000591424.1 |
GJC1 |
gap junction protein, gamma 1, 45kDa |
| chr18_+_3447572 | 0.54 |
ENST00000548489.2 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chr16_-_66583701 | 0.54 |
ENST00000527800.1 ENST00000525974.1 ENST00000563369.2 |
TK2 |
thymidine kinase 2, mitochondrial |
| chr11_-_118966167 | 0.54 |
ENST00000530167.1 |
H2AFX |
H2A histone family, member X |
| chr2_+_130737223 | 0.54 |
ENST00000410061.2 |
RAB6C |
RAB6C, member RAS oncogene family |
| chr2_+_161993412 | 0.53 |
ENST00000259075.2 ENST00000432002.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr5_+_72143988 | 0.53 |
ENST00000506351.2 |
TNPO1 |
transportin 1 |
| chr12_+_5541267 | 0.51 |
ENST00000423158.3 |
NTF3 |
neurotrophin 3 |
| chr5_-_157002749 | 0.49 |
ENST00000517905.1 ENST00000430702.2 ENST00000394020.1 |
ADAM19 |
ADAM metallopeptidase domain 19 |
| chr4_+_129730779 | 0.49 |
ENST00000226319.6 |
PHF17 |
jade family PHD finger 1 |
| chr11_+_65407331 | 0.49 |
ENST00000527525.1 |
SIPA1 |
signal-induced proliferation-associated 1 |
| chr8_-_145018080 | 0.48 |
ENST00000354589.3 |
PLEC |
plectin |
| chr17_-_19290483 | 0.47 |
ENST00000395592.2 ENST00000299610.4 |
MFAP4 |
microfibrillar-associated protein 4 |
| chr19_+_17326141 | 0.47 |
ENST00000445667.2 ENST00000263897.5 |
USE1 |
unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
| chr22_-_24641027 | 0.46 |
ENST00000398292.3 ENST00000263112.7 ENST00000418439.2 ENST00000424217.1 ENST00000327365.4 |
GGT5 |
gamma-glutamyltransferase 5 |
| chr14_+_79745682 | 0.46 |
ENST00000557594.1 |
NRXN3 |
neurexin 3 |
| chr9_-_115983568 | 0.44 |
ENST00000446284.1 ENST00000414250.1 |
FKBP15 |
FK506 binding protein 15, 133kDa |
| chr5_+_152870734 | 0.44 |
ENST00000521843.2 |
GRIA1 |
glutamate receptor, ionotropic, AMPA 1 |
| chr17_-_40333099 | 0.44 |
ENST00000607371.1 |
KCNH4 |
potassium voltage-gated channel, subfamily H (eag-related), member 4 |
| chr12_+_133757995 | 0.41 |
ENST00000536435.2 ENST00000228289.5 ENST00000541211.2 ENST00000500625.3 ENST00000539248.2 ENST00000542711.2 ENST00000536899.2 ENST00000542986.2 ENST00000537565.1 ENST00000541975.2 |
ZNF268 |
zinc finger protein 268 |
| chr17_+_48172639 | 0.40 |
ENST00000503176.1 ENST00000503614.1 |
PDK2 |
pyruvate dehydrogenase kinase, isozyme 2 |
| chr18_+_32558208 | 0.39 |
ENST00000436190.2 |
MAPRE2 |
microtubule-associated protein, RP/EB family, member 2 |
| chr1_+_207943667 | 0.39 |
ENST00000462968.2 |
CD46 |
CD46 molecule, complement regulatory protein |
| chr9_-_115983641 | 0.38 |
ENST00000238256.3 |
FKBP15 |
FK506 binding protein 15, 133kDa |
| chr10_-_32345305 | 0.38 |
ENST00000302418.4 |
KIF5B |
kinesin family member 5B |
| chr15_+_57210818 | 0.38 |
ENST00000438423.2 ENST00000267811.5 ENST00000452095.2 ENST00000559609.1 ENST00000333725.5 |
TCF12 |
transcription factor 12 |
| chr3_+_138340067 | 0.38 |
ENST00000479848.1 |
FAIM |
Fas apoptotic inhibitory molecule |
| chr16_+_56623433 | 0.35 |
ENST00000570176.1 |
MT3 |
metallothionein 3 |
| chr3_+_148457585 | 0.35 |
ENST00000402260.1 |
AGTR1 |
angiotensin II receptor, type 1 |
| chr4_-_74486347 | 0.32 |
ENST00000342081.3 |
RASSF6 |
Ras association (RalGDS/AF-6) domain family member 6 |
| chr12_+_64173583 | 0.32 |
ENST00000261234.6 |
TMEM5 |
transmembrane protein 5 |
| chr1_-_203320617 | 0.31 |
ENST00000354955.4 |
FMOD |
fibromodulin |
| chr20_-_17662705 | 0.31 |
ENST00000455029.2 |
RRBP1 |
ribosome binding protein 1 |
| chr2_+_204193129 | 0.30 |
ENST00000417864.1 |
ABI2 |
abl-interactor 2 |
| chr5_+_152870287 | 0.30 |
ENST00000340592.5 |
GRIA1 |
glutamate receptor, ionotropic, AMPA 1 |
| chr2_+_162016827 | 0.30 |
ENST00000429217.1 ENST00000406287.1 ENST00000402568.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr5_-_81574160 | 0.28 |
ENST00000510210.1 ENST00000512493.1 ENST00000507980.1 ENST00000511844.1 ENST00000510019.1 |
RPS23 |
ribosomal protein S23 |
| chr1_+_152658599 | 0.28 |
ENST00000368780.3 |
LCE2B |
late cornified envelope 2B |
| chr12_-_56710118 | 0.27 |
ENST00000273308.4 |
CNPY2 |
canopy FGF signaling regulator 2 |
| chr2_+_204193149 | 0.26 |
ENST00000422511.2 |
ABI2 |
abl-interactor 2 |
| chr1_+_153963227 | 0.25 |
ENST00000368567.4 ENST00000392558.4 |
RPS27 |
ribosomal protein S27 |
| chr17_+_47865917 | 0.24 |
ENST00000259021.4 ENST00000454930.2 ENST00000509773.1 ENST00000510819.1 ENST00000424009.2 |
KAT7 |
K(lysine) acetyltransferase 7 |
| chr1_+_16767195 | 0.23 |
ENST00000504551.2 ENST00000457722.2 ENST00000406746.1 ENST00000443980.2 |
NECAP2 |
NECAP endocytosis associated 2 |
| chr2_+_201994208 | 0.23 |
ENST00000440180.1 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
| chr4_+_71108300 | 0.22 |
ENST00000304954.3 |
CSN3 |
casein kappa |
| chr5_-_9630463 | 0.22 |
ENST00000382492.2 |
TAS2R1 |
taste receptor, type 2, member 1 |
| chr3_+_183353356 | 0.21 |
ENST00000242810.6 ENST00000493074.1 ENST00000437402.1 ENST00000454495.2 ENST00000473045.1 ENST00000468101.1 ENST00000427201.2 ENST00000482138.1 ENST00000454652.2 |
KLHL24 |
kelch-like family member 24 |
| chr5_-_114598548 | 0.19 |
ENST00000379615.3 ENST00000419445.1 |
PGGT1B |
protein geranylgeranyltransferase type I, beta subunit |
| chr1_+_22333943 | 0.19 |
ENST00000400271.2 |
CELA3A |
chymotrypsin-like elastase family, member 3A |
| chr5_-_78281603 | 0.19 |
ENST00000264914.4 |
ARSB |
arylsulfatase B |
| chr1_+_178995021 | 0.18 |
ENST00000263733.4 |
FAM20B |
family with sequence similarity 20, member B |
| chr2_+_161993465 | 0.18 |
ENST00000457476.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr11_+_57365150 | 0.18 |
ENST00000457869.1 ENST00000340687.6 ENST00000378323.4 ENST00000378324.2 ENST00000403558.1 |
SERPING1 |
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr16_+_4674787 | 0.18 |
ENST00000262370.7 |
MGRN1 |
mahogunin ring finger 1, E3 ubiquitin protein ligase |
| chr8_-_7309887 | 0.17 |
ENST00000458665.1 ENST00000528168.1 |
SPAG11B |
sperm associated antigen 11B |
| chr7_-_137686791 | 0.16 |
ENST00000452463.1 ENST00000330387.6 ENST00000456390.1 |
CREB3L2 |
cAMP responsive element binding protein 3-like 2 |
| chr9_+_115983808 | 0.16 |
ENST00000374210.6 ENST00000374212.4 |
SLC31A1 |
solute carrier family 31 (copper transporter), member 1 |
| chr8_-_101734170 | 0.16 |
ENST00000522387.1 ENST00000518196.1 |
PABPC1 |
poly(A) binding protein, cytoplasmic 1 |
| chr3_-_10362725 | 0.16 |
ENST00000397109.3 ENST00000428626.1 ENST00000445064.1 ENST00000431352.1 ENST00000397117.1 ENST00000337354.4 ENST00000383801.2 ENST00000432213.1 ENST00000350697.3 |
SEC13 |
SEC13 homolog (S. cerevisiae) |
| chr12_-_57081940 | 0.16 |
ENST00000436399.2 |
PTGES3 |
prostaglandin E synthase 3 (cytosolic) |
| chr8_-_141774467 | 0.16 |
ENST00000520151.1 ENST00000519024.1 ENST00000519465.1 |
PTK2 |
protein tyrosine kinase 2 |
| chr2_+_172378757 | 0.16 |
ENST00000409484.1 ENST00000321348.4 ENST00000375252.3 |
CYBRD1 |
cytochrome b reductase 1 |
| chr4_-_13546632 | 0.15 |
ENST00000382438.5 |
NKX3-2 |
NK3 homeobox 2 |
| chr3_+_138340049 | 0.14 |
ENST00000464668.1 |
FAIM |
Fas apoptotic inhibitory molecule |
| chr12_+_103981044 | 0.14 |
ENST00000388887.2 |
STAB2 |
stabilin 2 |
| chr19_-_46105411 | 0.14 |
ENST00000323040.4 ENST00000544371.1 |
GPR4 OPA3 |
G protein-coupled receptor 4 optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia) |
| chr1_+_22351977 | 0.14 |
ENST00000420503.1 ENST00000416769.1 ENST00000404210.2 |
LINC00339 |
long intergenic non-protein coding RNA 339 |
| chr14_+_79746249 | 0.12 |
ENST00000428277.2 |
NRXN3 |
neurexin 3 |
| chr3_+_46412345 | 0.12 |
ENST00000292303.4 |
CCR5 |
chemokine (C-C motif) receptor 5 (gene/pseudogene) |
| chr3_+_184033135 | 0.11 |
ENST00000424196.1 |
EIF4G1 |
eukaryotic translation initiation factor 4 gamma, 1 |
| chr1_-_157746909 | 0.11 |
ENST00000392274.3 ENST00000361516.3 ENST00000368181.4 |
FCRL2 |
Fc receptor-like 2 |
| chr5_-_178054105 | 0.10 |
ENST00000316308.4 |
CLK4 |
CDC-like kinase 4 |
| chr15_+_57210961 | 0.10 |
ENST00000557843.1 |
TCF12 |
transcription factor 12 |
| chr17_-_4852332 | 0.10 |
ENST00000572383.1 |
PFN1 |
profilin 1 |
| chr17_-_4852243 | 0.10 |
ENST00000225655.5 |
PFN1 |
profilin 1 |
Gene Ontology Analysis
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.8 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.2 | 2.8 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.2 | 5.4 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.1 | 2.3 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 2.0 | REACTOME SIGNALING BY NOTCH2 | Genes involved in Signaling by NOTCH2 |
| 0.1 | 3.8 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 6.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 4.1 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.1 | 2.0 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 1.8 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 1.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 2.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 1.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 3.0 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 2.3 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 2.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.5 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 3.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.6 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.7 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 1.3 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 2.4 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 1.0 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 1.0 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.4 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 1.2 | REACTOME IL 3 5 AND GM CSF SIGNALING | Genes involved in Interleukin-3, 5 and GM-CSF signaling |
| 0.0 | 0.0 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.7 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 1.3 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.5 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 7.8 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 1.6 | 28.0 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.8 | 2.4 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.6 | 5.4 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.6 | 8.3 | GO:0033643 | host cell part(GO:0033643) |
| 0.5 | 5.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.4 | 2.8 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.4 | 6.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.2 | 0.9 | GO:0043260 | laminin-10 complex(GO:0043259) laminin-11 complex(GO:0043260) |
| 0.2 | 1.3 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.2 | 3.4 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.2 | 3.0 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.2 | 2.0 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 0.7 | GO:0044308 | axonal spine(GO:0044308) |
| 0.1 | 1.4 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 0.4 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.1 | 10.4 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.1 | 1.4 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 1.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 0.4 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 11.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 2.9 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 2.9 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 3.2 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 1.4 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.1 | 8.1 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 1.0 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 0.2 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.1 | 4.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 0.4 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 8.7 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 0.4 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.1 | 0.8 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 1.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.8 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 1.0 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 1.2 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.5 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.8 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 1.4 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.6 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 4.4 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 1.5 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 17.1 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 3.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.8 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.5 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 0.2 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 15.1 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.3 | 28.0 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.2 | 10.0 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.2 | 7.1 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 6.8 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 3.6 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 5.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 4.2 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 2.0 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 2.9 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 18.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 8.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 0.9 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 2.1 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 1.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 3.7 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 3.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 3.6 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 1.2 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 1.2 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 2.2 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.7 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 1.3 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.5 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.4 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.5 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.2 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.3 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 10.6 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 1.9 | 7.8 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 1.6 | 4.9 | GO:0018262 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 1.5 | 4.6 | GO:1990523 | bone regeneration(GO:1990523) |
| 1.5 | 11.7 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 1.4 | 8.3 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 1.3 | 5.4 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 1.1 | 3.4 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 1.1 | 6.7 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.8 | 3.3 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.8 | 28.0 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.7 | 5.2 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.7 | 2.2 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.6 | 2.8 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.5 | 4.7 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.5 | 2.1 | GO:1905123 | regulation of endosome organization(GO:1904978) regulation of glucosylceramidase activity(GO:1905123) |
| 0.5 | 4.1 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.5 | 1.8 | GO:0046104 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.4 | 2.2 | GO:0045914 | negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) regulation of peroxidase activity(GO:2000468) positive regulation of peroxidase activity(GO:2000470) |
| 0.4 | 9.5 | GO:1901881 | positive regulation of protein depolymerization(GO:1901881) |
| 0.4 | 2.4 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.4 | 2.3 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.4 | 3.3 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.4 | 2.1 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) regulation of nodal signaling pathway(GO:1900107) |
| 0.3 | 6.2 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.3 | 2.3 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.3 | 2.6 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.3 | 2.3 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.3 | 3.6 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.3 | 1.5 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.3 | 2.1 | GO:0019374 | galactosylceramide metabolic process(GO:0006681) galactolipid metabolic process(GO:0019374) |
| 0.2 | 1.0 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) positive regulation of potassium ion import(GO:1903288) |
| 0.2 | 1.6 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.2 | 2.7 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.2 | 8.0 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.2 | 5.3 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.1 | 0.6 | GO:0086053 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 0.1 | 0.4 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.1 | 0.8 | GO:0072564 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.1 | 1.3 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.1 | 1.6 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 0.8 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.1 | 1.4 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.5 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.1 | 0.3 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.1 | 2.0 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.1 | 1.2 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.1 | 0.3 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.1 | 1.2 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) limb bud formation(GO:0060174) |
| 0.1 | 1.4 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 3.6 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 2.3 | GO:0007567 | parturition(GO:0007567) |
| 0.1 | 0.7 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.1 | 0.9 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.4 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.1 | 1.3 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.1 | 0.2 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.1 | 0.2 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.1 | 0.9 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.1 | 2.6 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.1 | 3.8 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 0.8 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.1 | 1.8 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.1 | 3.0 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.5 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 1.8 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 0.8 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 1.4 | GO:0035966 | response to unfolded protein(GO:0006986) response to topologically incorrect protein(GO:0035966) |
| 0.0 | 2.9 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 3.5 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 1.2 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 1.1 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 1.0 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 1.2 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 1.7 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 2.5 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.4 | GO:0043382 | positive regulation of memory T cell differentiation(GO:0043382) |
| 0.0 | 1.0 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 1.1 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.5 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0072141 | renal interstitial fibroblast development(GO:0072141) |
| 0.0 | 0.5 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 1.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.5 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.2 | GO:0015677 | copper ion import(GO:0015677) |
| 0.0 | 3.6 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.0 | 0.2 | GO:0010822 | positive regulation of mitochondrion organization(GO:0010822) |
| 0.0 | 0.7 | GO:0098779 | mitophagy in response to mitochondrial depolarization(GO:0098779) |
| 0.0 | 0.3 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.0 | 0.3 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.2 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.1 | GO:0019064 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.2 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.2 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.4 | GO:0050812 | regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.5 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.2 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.0 | 7.1 | GO:0006260 | DNA replication(GO:0006260) |
| 0.0 | 0.2 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.1 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.1 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.5 | GO:0010824 | regulation of centrosome duplication(GO:0010824) |
| 0.0 | 1.5 | GO:0007041 | lysosomal transport(GO:0007041) |
| 0.0 | 1.2 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.0 | 0.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.3 | GO:0018126 | protein hydroxylation(GO:0018126) |
| 0.0 | 0.2 | GO:0007635 | chemosensory behavior(GO:0007635) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 28.0 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 1.8 | 5.4 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 1.7 | 5.2 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 1.3 | 10.6 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 1.3 | 11.7 | GO:0048495 | Roundabout binding(GO:0048495) |
| 1.1 | 3.3 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.8 | 3.8 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.8 | 5.3 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.7 | 4.3 | GO:0004471 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) oxaloacetate decarboxylase activity(GO:0008948) |
| 0.7 | 3.6 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.6 | 9.5 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.6 | 4.9 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.6 | 1.8 | GO:0004797 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.5 | 8.3 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.5 | 2.3 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.4 | 2.2 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.4 | 2.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.4 | 3.4 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.4 | 6.7 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.4 | 2.3 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.3 | 3.3 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.3 | 2.9 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.3 | 1.2 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.3 | 2.4 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.2 | 0.4 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) protein histidine kinase activity(GO:0004673) |
| 0.2 | 5.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 2.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.6 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 0.1 | 1.4 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 0.8 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 0.5 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.1 | 4.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.3 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.1 | 2.8 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 3.8 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 0.4 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 2.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 1.6 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 0.4 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.1 | 1.3 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 0.7 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 3.6 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 1.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.2 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.1 | 6.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 2.8 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 1.6 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 8.5 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.8 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.9 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 3.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0000982 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) |
| 0.0 | 3.0 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 1.5 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.5 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.2 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 4.0 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.6 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 2.2 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.4 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.0 | 0.2 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.5 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.7 | GO:0004950 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 1.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.7 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.4 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.0 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.1 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |