|
chr6_-_41362370
|
1.285
|
NM_018643
|
TREM1
|
triggering receptor expressed on myeloid cells 1
|
|
chr2_+_132196533
|
1.139
|
NM_013310
|
C2orf27A
|
chromosome 2 open reading frame 27A
|
|
chrX_-_131089552
|
0.980
|
NM_194277
|
FRMD7
|
FERM domain containing 7
|
|
chr10_-_23673777
|
0.972
|
NM_153714
|
C10orf67
|
chromosome 10 open reading frame 67
|
|
chr3_+_63238854
|
0.914
|
NM_001130003
|
SYNPR
|
synaptoporin
|
|
chr2_+_204279442
|
0.910
|
NM_006139
|
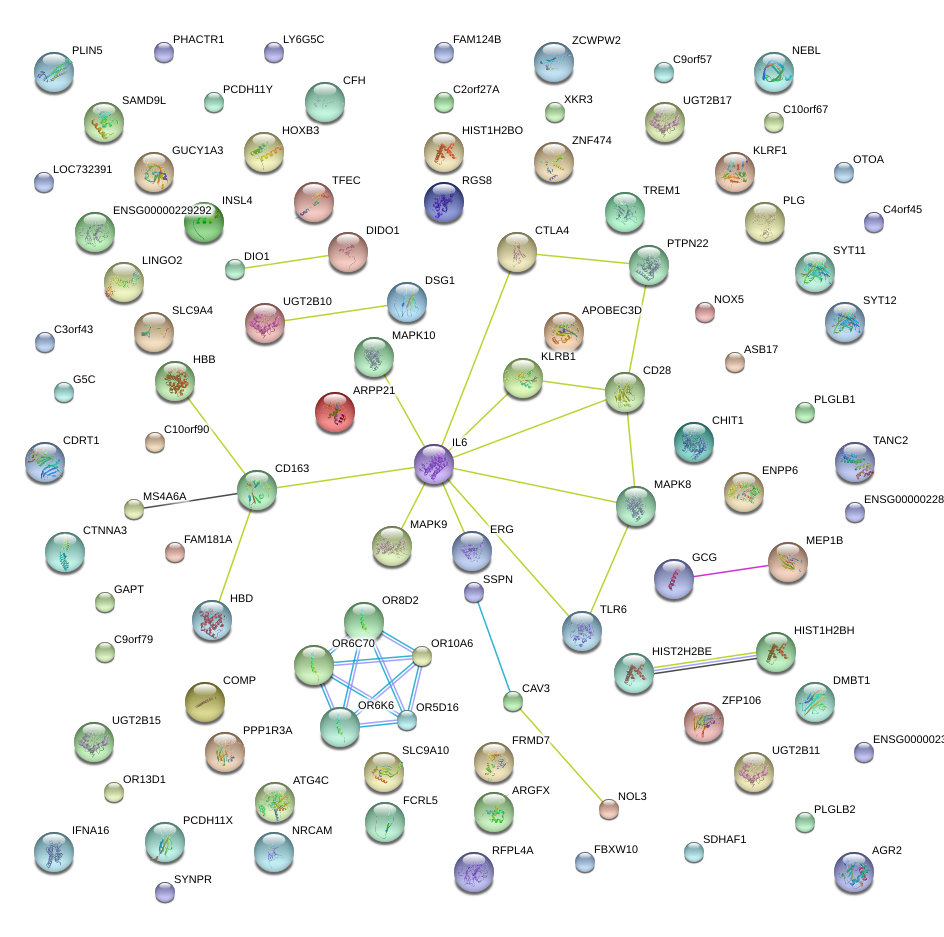
CD28
|
CD28 molecule
|
|
chr9_+_89687560
|
0.895
|
NM_178828
|
C9orf79
|
chromosome 9 open reading frame 79
|
|
chr11_-_5212168
|
0.880
|
NM_000519
|
HBD
|
hemoglobin, delta
|
|
chr7_-_115458010
|
0.854
|
NM_001018058
NM_012252
|
TFEC
|
transcription factor EC
|
|
chr10_-_42183441
|
0.849
|
|
LOC283027
|
hypothetical protein LOC283027
|
|
chr7_-_107667728
|
0.845
|
NM_001037132
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr3_+_122769467
|
0.843
|
NM_001012659
|
ARGFX
|
arginine-fifty homeobox
|
|
chr1_+_559129
|
0.820
|
|
|
|
|
chr19_+_53450743
|
0.808
|
|
|
|
|
chr3_+_8750485
|
0.807
|
NM_033337
NM_001234
|
CAV3
|
caveolin 3
|
|
chr6_+_12825818
|
0.747
|
NM_030948
|
PHACTR1
|
phosphatase and actin regulator 1
|
|
chr11_+_55362802
|
0.739
|
NM_001005496
|
OR5D16
|
olfactory receptor, family 5, subfamily D, member 16
|
|
chr7_+_22733290
|
0.731
|
NM_000600
|
IL6
|
interleukin 6 (interferon, beta 2)
|
|
chr3_-_113495763
|
0.700
|
NM_183061
|
SLC9A10
|
solute carrier family 9, member 10
|
|
chr10_+_49279647
|
0.699
|
NM_002750
NM_139046
NM_139047
NM_139049
|
MAPK8
|
mitogen-activated protein kinase 8
|
|
chr9_-_21207303
|
0.696
|
NM_002173
|
IFNA16
|
interferon, alpha 16
|
|
chr1_-_114215768
|
0.683
|
NM_001193431
NM_012411
NM_015967
|
PTPN22
|
protein tyrosine phosphatase, non-receptor type 22 (lymphoid)
|
|
chr10_+_124310170
|
0.681
|
NM_004406
NM_007329
NM_017579
|
DMBT1
|
deleted in malignant brain tumors 1
|
|
chr17_-_44006808
|
0.675
|
NM_002146
|
HOXB3
|
homeobox B3
|
|
chr4_+_156807311
|
0.672
|
NM_000856
NM_001130682
NM_001130686
NM_001130687
NM_001130683
NM_001130685
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3
|
|
chr1_+_54132447
|
0.669
|
NM_000792
NM_001039715
NM_001039716
NM_213593
|
DIO1
|
deiodinase, iodothyronine, type I
|
|
chr1_+_156991133
|
0.662
|
NM_001005184
|
OR6K6
|
olfactory receptor, family 6, subfamily K, member 6
|
|
chr10_-_21226536
|
0.653
|
NM_006393
|
NEBL
|
nebulette
|
|
chr19_+_41177882
|
0.651
|
NM_001042631
|
SDHAF1
|
succinate dehydrogenase complex assembly factor 1
|
|
chr22_-_15682555
|
0.648
|
NM_175878
|
XKR3
|
XK, Kell blood group complex subunit-related family, member 3
|
|
chr19_-_21438525
|
0.642
|
|
|
|
|
chr3_+_28406990
|
0.641
|
NM_001040432
|
ZCWPW2
|
zinc finger, CW type with PWWP domain 2
|
|
chr1_+_180685878
|
0.634
|
NM_001137669
|
RGSL1
|
regulator of G-protein signaling like 1
|
|
chr1_-_155788775
|
0.634
|
NM_001195388
NM_031281
|
FCRL5
|
Fc receptor-like 5
|
|
chr4_-_38534802
|
0.634
|
NM_006068
|
TLR6
|
toll-like receptor 6
|
|
chr2_-_162717159
|
0.627
|
NM_002054
|
GCG
|
glucagon
|
|
chr5_+_121493113
|
0.623
|
NM_207317
|
ZNF474
|
zinc finger protein 474
|
|
chr1_+_559618
|
0.620
|
|
|
|
|
chr12_+_26239535
|
0.617
|
NM_001135823
|
SSPN
|
sarcospan (Kras oncogene-associated gene)
|
|
chr9_-_73865318
|
0.614
|
NM_001128618
|
C9orf57
|
chromosome 9 open reading frame 57
|
|
chr5_+_57823017
|
0.606
|
NM_152687
|
GAPT
|
GRB2-binding adaptor protein, transmembrane
|
|
chr21_-_38792173
|
0.603
|
NM_001136155
NM_182918
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr12_-_7547552
|
0.592
|
NM_004244
NM_203416
|
CD163
|
CD163 molecule
|
|
chr1_-_180908639
|
0.590
|
NM_001102450
NM_033345
|
RGS8
|
regulator of G-protein signaling 8
|
|
chr7_-_113346284
|
0.586
|
NM_002711
|
PPP1R3A
|
protein phosphatase 1, regulatory (inhibitor) subunit 3A
|
|
chr10_-_128199986
|
0.581
|
NM_001004298
|
C10orf90
|
chromosome 10 open reading frame 90
|
|
chr12_+_9871343
|
0.573
|
NM_016523
|
KLRF1
|
killer cell lectin-like receptor subfamily F, member 1
|
|
chr1_+_194887630
|
0.566
|
NM_000186
NM_001014975
|
CFH
|
complement factor H
|
|
chr14_+_93455010
|
0.556
|
NM_138344
|
FAM181A
|
family with sequence similarity 181, member A
|
|
chr2_-_224974954
|
0.555
|
NM_001122779
NM_024785
|
FAM124B
|
family with sequence similarity 124B
|
|
chr6_-_31756119
|
0.550
|
NM_025262
|
LY6G5C
|
lymphocyte antigen 6 complex, locus G5C
|
|
chr16_+_21603250
|
0.544
|
NM_001161683
|
OTOA
|
otoancorin
|
|
chr4_-_185376012
|
0.542
|
NM_153343
|
ENPP6
|
ectonucleotide pyrophosphatase/phosphodiesterase 6
|
|
chr4_-_69218909
|
0.539
|
NM_001076
|
UGT2B15
|
UDP glucuronosyltransferase 2 family, polypeptide B15
|
|
chr7_-_16811131
|
0.536
|
NM_006408
|
AGR2
|
anterior gradient homolog 2 (Xenopus laevis)
|
|
chr11_-_7906784
|
0.532
|
NM_001004461
|
OR10A6
|
olfactory receptor, family 10, subfamily A, member 6
|
|
chr7_-_112545847
|
0.530
|
|
LOC401397
|
hypothetical LOC401397
|
|
chr17_+_18588050
|
0.526
|
NM_031456
|
FBXW10
CDRT1
|
F-box and WD repeat domain containing 10
CMT1A duplicated region transcript 1
|
|
chr1_-_76170563
|
0.512
|
NM_080868
|
ASB17
|
ankyrin repeat and SOCS box containing 17
|
|
chr19_+_60962318
|
0.505
|
NM_001145014
|
RFPL4A
|
ret finger protein-like 4A
|
|
chr9_-_28709302
|
0.505
|
NM_152570
|
LINGO2
|
leucine rich repeat and Ig domain containing 2
|
|
chr2_-_87102453
|
0.503
|
NM_002665
NM_001032392
|
PLGLB2
PLGLB1
|
plasminogen-like B2
plasminogen-like B1
|
|
chr9_+_5221401
|
0.498
|
NM_002195
|
INSL4
|
insulin-like 4 (placenta)
|
|
chr4_-_69570919
|
0.497
|
NM_001076
|
UGT2B15
|
UDP glucuronosyltransferase 2 family, polypeptide B15
|
|
chr2_+_102456193
|
0.493
|
NM_001011552
|
SLC9A4
|
solute carrier family 9 (sodium/hydrogen exchanger), member 4
|
|
chr15_+_67094087
|
0.493
|
NM_001184779
NM_024505
|
NOX5
|
NADPH oxidase, EF-hand calcium binding domain 5
|
|
chr6_+_26359805
|
0.487
|
NM_003524
|
HIST1H2BH
|
histone cluster 1, H2bh
|
|
chr12_-_9651748
|
0.484
|
NM_002258
|
KLRB1
|
killer cell lectin-like receptor subfamily B, member 1
|
|
chr4_-_160175782
|
0.475
|
NM_152543
|
C4orf45
|
chromosome 4 open reading frame 45
|
|
chr3_-_197726587
|
0.474
|
NM_001077657
|
C3orf43
|
chromosome 3 open reading frame 43
|
|
chr1_+_154120170
|
0.469
|
|
SYT11
|
synaptotagmin XI
|
|
chr3_+_35658852
|
0.461
|
NM_016300
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr12_-_54150188
|
0.457
|
NM_001005499
|
OR6C70
|
olfactory receptor, family 6, subfamily C, member 70
|
|
chr7_-_92615593
|
0.453
|
NM_152703
|
SAMD9L
|
sterile alpha motif domain containing 9-like
|
|
chr11_-_59707074
|
0.451
|
NM_022349
NM_152851
NM_152852
|
MS4A6A
|
membrane-spanning 4-domains, subfamily A, member 6A
|
|
chr19_-_18763077
|
0.450
|
NM_000095
|
COMP
|
cartilage oligomeric matrix protein
|
|
chr10_-_69095362
|
0.442
|
NM_001127384
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3
|
|
chr1_+_63022359
|
0.442
|
NM_032852
NM_178221
|
ATG4C
|
ATG4 autophagy related 4 homolog C (S. cerevisiae)
|
|
chr1_-_201465395
|
0.438
|
NM_003465
|
CHIT1
|
chitinase 1 (chitotriosidase)
|
|
chr11_-_123695302
|
0.437
|
NM_001002918
|
OR8D2
|
olfactory receptor, family 8, subfamily D, member 2
|
|
chr9_+_106496480
|
0.434
|
NM_001004484
|
OR13D1
|
olfactory receptor, family 13, subfamily D, member 1
|
|
chr19_-_4486202
|
0.432
|
NM_001013706
|
PLIN5
|
perilipin 5
|
|
chr17_+_58440629
|
0.415
|
NM_025185
|
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2
|
|
chr18_+_28023984
|
0.408
|
NM_005925
|
MEP1B
|
meprin A, beta
|
|
chr18_+_27152049
|
0.402
|
NM_001942
|
DSG1
|
desmoglein 1
|
|
chr5_+_76470829
|
0.402
|
|
|
|
|
chrX_+_90976314
|
0.398
|
NM_014522
NM_032968
NM_032969
|
PCDH11X
|
protocadherin 11 X-linked
|
|
chr7_-_81237162
|
0.395
|
NM_000601
NM_001010931
NM_001010932
NM_001010933
NM_001010934
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor)
|
|
chrX_+_90977115
|
0.392
|
NM_001168360
NM_001168361
NM_001168362
NM_001168363
|
PCDH11X
|
protocadherin 11 X-linked
|
|
chr3_+_110338250
|
0.386
|
|
FLJ22763
|
hypothetical LOC401081
|
|
chr18_-_3835295
|
0.384
|
NM_001003809
|
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1
|
|
chr5_-_176258920
|
0.381
|
NM_002115
|
HK3
|
hexokinase 3 (white cell)
|
|
chr3_+_63613383
|
0.381
|
NM_001080537
|
SNTN
|
sentan, cilia apical structure protein
|
|
chr12_-_61832856
|
0.374
|
NM_000706
|
AVPR1A
|
arginine vasopressin receptor 1A
|
|
chr7_+_143043871
|
0.373
|
NM_001130026
|
FAM115C
|
family with sequence similarity 115, member C
|
|
chr19_-_6384772
|
0.370
|
NM_173637
|
SLC25A41
|
solute carrier family 25, member 41
|
|
chr4_+_120276386
|
0.368
|
NM_016599
|
MYOZ2
|
myozenin 2
|
|
chr22_-_41815277
|
0.365
|
NM_012263
|
TTLL1
|
tubulin tyrosine ligase-like family, member 1
|
|
chr2_+_204279624
|
0.364
|
|
CD28
|
CD28 molecule
|
|
chrX_+_66705407
|
0.362
|
NM_001011645
|
AR
|
androgen receptor
|
|
chr3_+_44667504
|
0.361
|
|
ZNF35
|
zinc finger protein 35
|
|
chr22_+_23532135
|
0.361
|
NM_001039948
NM_001098497
NM_001098498
NM_133454
|
SGSM1
|
small G protein signaling modulator 1
|
|
chrX_+_154764136
|
0.361
|
NM_001145149
NM_001185183
NM_005638
|
VAMP7
|
vesicle-associated membrane protein 7
|
|
chr22_-_22252376
|
0.356
|
NM_020070
NM_152855
|
IGLL1
|
immunoglobulin lambda-like polypeptide 1
|
|
chr6_+_31791063
|
0.355
|
NM_021246
|
LY6G6D
|
lymphocyte antigen 6 complex, locus G6D
|
|
chr15_-_88023576
|
0.353
|
NM_001145311
NM_002666
|
PLIN1
|
perilipin 1
|
|
chr1_-_67039518
|
0.349
|
NM_005478
|
INSL5
|
insulin-like 5
|
|
chr5_+_140594121
|
0.342
|
|
PCDHB18
|
protocadherin beta 18 pseudogene
|
|
chr11_-_51269023
|
0.339
|
NM_001005272
|
OR4A5
|
olfactory receptor, family 4, subfamily A, member 5
|
|
chr1_-_156954528
|
0.339
|
NM_001005327
|
OR6K3
|
olfactory receptor, family 6, subfamily K, member 3
|
|
chr7_+_150319076
|
0.338
|
NM_000603
|
NOS3
|
nitric oxide synthase 3 (endothelial cell)
|
|
chr3_-_168853982
|
0.336
|
NM_178824
|
WDR49
|
WD repeat domain 49
|
|
chr1_-_156923111
|
0.335
|
NM_003126
|
SPTA1
|
spectrin, alpha, erythrocytic 1 (elliptocytosis 2)
|
|
chr12_+_100512877
|
0.335
|
NM_002465
NM_206819
NM_206820
NM_206821
|
MYBPC1
|
myosin binding protein C, slow type
|
|
chr4_+_71713324
|
0.334
|
NM_031889
|
ENAM
|
enamelin
|
|
chr14_-_106084085
|
0.333
|
|
IGHV4-31
IGHG1
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr17_+_9670105
|
0.331
|
NM_004246
|
GLP2R
|
glucagon-like peptide 2 receptor
|
|
chr5_+_140200995
|
0.330
|
NM_018911
NM_031856
|
PCDHA8
|
protocadherin alpha 8
|
|
chr5_+_40877053
|
0.330
|
NM_032587
|
CARD6
|
caspase recruitment domain family, member 6
|
|
chr11_+_93940121
|
0.329
|
NM_152431
|
PIWIL4
|
piwi-like 4 (Drosophila)
|
|
chr5_+_140583098
|
0.329
|
NM_018934
|
PCDHB14
|
protocadherin beta 14
|
|
chr11_-_113971681
|
0.323
|
NM_001077639
NM_017678
|
FAM55D
|
family with sequence similarity 55, member D
|
|
chr3_+_150065732
|
0.323
|
NM_001870
|
CPA3
|
carboxypeptidase A3 (mast cell)
|
|
chr11_+_113351006
|
0.320
|
NM_000869
NM_213621
|
HTR3A
|
5-hydroxytryptamine (serotonin) receptor 3A
|
|
chr3_-_170347019
|
0.318
|
NM_001105078
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr1_-_156635879
|
0.317
|
NM_001004475
|
OR10T2
|
olfactory receptor, family 10, subfamily T, member 2
|
|
chr7_-_102070473
|
0.316
|
NM_001114403
|
UPK3BL
POLR2J2
|
uroplakin 3B-like
polymerase (RNA) II (DNA directed) polypeptide J2
|
|
chr7_+_37689903
|
0.314
|
|
|
|
|
chr3_-_46224791
|
0.307
|
NM_001295
|
CCR1
|
chemokine (C-C motif) receptor 1
|
|
chr3_+_49210864
|
0.306
|
NM_178173
|
CCDC36
|
coiled-coil domain containing 36
|
|
chr3_-_170864111
|
0.303
|
NM_004991
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr11_-_5321329
|
0.301
|
NM_001005567
|
OR51B5
|
olfactory receptor, family 51, subfamily B, member 5
|
|
chr5_+_140227951
|
0.300
|
NM_018902
NM_031861
|
PCDHA11
|
protocadherin alpha 11
|
|
chr7_-_86433139
|
0.299
|
NM_152748
|
KIAA1324L
|
KIAA1324-like
|
|
chr6_+_47857733
|
0.298
|
NM_181744
|
OPN5
|
opsin 5
|
|
chr5_-_39306465
|
0.294
|
|
FYB
|
FYN binding protein
|
|
chr11_-_76800525
|
0.294
|
|
PAK1
|
p21 protein (Cdc42/Rac)-activated kinase 1
|
|
chr10_+_118946989
|
0.292
|
NM_181840
|
KCNK18
|
potassium channel, subfamily K, member 18
|
|
chr17_+_25292748
|
0.289
|
NM_198529
|
EFCAB5
|
EF-hand calcium binding domain 5
|
|
chr8_-_111056099
|
0.285
|
NM_014379
|
KCNV1
|
potassium channel, subfamily V, member 1
|
|
chr4_+_43185
|
0.285
|
NM_182524
|
ZNF595
ZNF718
|
zinc finger protein 595
zinc finger protein 718
|
|
chr2_-_190635566
|
0.285
|
NM_005259
|
MSTN
|
myostatin
|
|
chr6_+_118976134
|
0.283
|
NM_002667
|
PLN
|
phospholamban
|
|
chr9_+_124416768
|
0.283
|
NM_012364
|
OR1Q1
|
olfactory receptor, family 1, subfamily Q, member 1
|
|
chr11_+_88550687
|
0.282
|
NM_000372
|
TYR
|
tyrosinase (oculocutaneous albinism IA)
|
|
chr2_+_87828718
|
0.281
|
NM_002665
NM_001032392
|
PLGLB2
PLGLB1
|
plasminogen-like B2
plasminogen-like B1
|
|
chr1_-_35143570
|
0.281
|
NM_001080418
|
DLGAP3
|
discs, large (Drosophila) homolog-associated protein 3
|
|
chr7_-_112545840
|
0.279
|
|
LOC401397
|
hypothetical LOC401397
|
|
chr20_+_34019942
|
0.278
|
NM_080834
|
C20orf152
|
chromosome 20 open reading frame 152
|
|
chr10_-_28611020
|
0.278
|
NM_173496
|
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7)
|
|
chr5_+_147562531
|
0.277
|
NM_001195290
NM_205841
|
SPINK6
|
serine peptidase inhibitor, Kazal type 6
|
|
chr5_-_35083996
|
0.275
|
NM_031900
|
AGXT2
|
alanine--glyoxylate aminotransferase 2
|
|
chr11_+_120478584
|
0.274
|
NM_005422
|
TECTA
|
tectorin alpha
|
|
chr13_+_22653059
|
0.273
|
NM_000231
|
SGCG
|
sarcoglycan, gamma (35kDa dystrophin-associated glycoprotein)
|
|
chr5_-_14706486
|
0.271
|
|
|
|
|
chr19_-_4510713
|
0.270
|
|
SEMA6B
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6B
|
|
chr10_+_69539255
|
0.264
|
NM_032578
|
MYPN
|
myopalladin
|
|
chr1_+_157676135
|
0.264
|
NM_012351
|
OR10J1
|
olfactory receptor, family 10, subfamily J, member 1
|
|
chr18_-_54447168
|
0.264
|
NM_052947
|
ALPK2
|
alpha-kinase 2
|
|
chr11_+_123600553
|
0.264
|
NM_001007249
|
OR8G2
|
olfactory receptor, family 8, subfamily G, member 2
|
|
chr22_-_38619739
|
0.260
|
NM_152512
|
ENTHD1
|
ENTH domain containing 1
|
|
chr1_+_156590109
|
0.258
|
NM_001042583
NM_001042584
NM_001042585
NM_001042586
NM_001042587
NM_001185107
NM_001185108
NM_001185110
NM_001185112
NM_001185113
NM_001185114
NM_001185115
NM_030893
|
CD1E
|
CD1e molecule
|
|
chr6_-_50097606
|
0.257
|
NM_001037497
NM_001037728
|
DEFB110
|
defensin, beta 110 locus
|
|
chr4_-_26100986
|
0.257
|
NM_000730
|
CCKAR
|
cholecystokinin A receptor
|
|
chr5_+_140605100
|
0.257
|
|
PCDHB15
|
protocadherin beta 15
|
|
chr19_-_5821546
|
0.256
|
NM_002034
|
FUT5
|
fucosyltransferase 5 (alpha (1,3) fucosyltransferase)
|
|
chr22_-_14829803
|
0.254
|
NM_001005239
|
OR11H1
|
olfactory receptor, family 11, subfamily H, member 1
|
|
chr7_-_15568099
|
0.254
|
NM_001004320
|
AGMO
|
alkylglycerol monooxygenase
|
|
chr3_+_123817151
|
0.254
|
NM_152615
|
PARP15
|
poly (ADP-ribose) polymerase family, member 15
|
|
chr3_+_69895310
|
0.253
|
NM_001184967
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr12_+_53177789
|
0.251
|
|
NCKAP1L
|
NCK-associated protein 1-like
|
|
chr2_+_102320148
|
0.251
|
NM_003856
|
IL1RL1
|
interleukin 1 receptor-like 1
|
|
chr5_-_158690047
|
0.250
|
NM_002187
|
IL12B
|
interleukin 12B (natural killer cell stimulatory factor 2, cytotoxic lymphocyte maturation factor 2, p40)
|
|
chr19_+_12064077
|
0.250
|
|
ZNF788
|
zinc finger family member 788
|
|
chr7_+_73826244
|
0.246
|
NM_000265
|
NCF1
NCF1B
NCF1C
|
neutrophil cytosolic factor 1
neutrophil cytosolic factor 1B pseudogene
neutrophil cytosolic factor 1C pseudogene
|
|
chr12_-_69434598
|
0.246
|
NM_130846
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R
|
|
chr12_+_20739555
|
0.244
|
NM_001145945
NM_001145946
NM_001145944
NM_017435
|
SLCO1C1
|
solute carrier organic anion transporter family, member 1C1
|
|
chr9_+_101628829
|
0.244
|
NM_173200
|
NR4A3
|
nuclear receptor subfamily 4, group A, member 3
|
|
chr19_+_52908412
|
0.243
|
NM_014601
|
EHD2
|
EH-domain containing 2
|
|
chr12_+_53177758
|
0.242
|
NM_005337
|
NCKAP1L
|
NCK-associated protein 1-like
|
|
chr4_-_123761615
|
0.241
|
NM_021803
|
IL21
|
interleukin 21
|
|
chr4_+_74566325
|
0.240
|
NM_001133
|
AFM
|
afamin
|
|
chr13_+_107719977
|
0.240
|
NM_001145645
NM_006573
|
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b
|
|
chr13_+_50813168
|
0.239
|
NM_001101320
|
SERPINE3
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 3
|
|
chr6_-_25982418
|
0.239
|
NM_001098486
NM_006632
|
SLC17A3
|
solute carrier family 17 (sodium phosphate), member 3
|
|
chr6_-_128281434
|
0.236
|
NM_001164687
|
THEMIS
|
thymocyte selection associated
|
|
chr3_-_9895633
|
0.235
|
NM_022094
|
CIDEC
|
cell death-inducing DFFA-like effector c
|
|
chr5_-_22889487
|
0.235
|
NM_004061
|
CDH12
|
cadherin 12, type 2 (N-cadherin 2)
|
|
chrX_-_23835868
|
0.235
|
NM_024122
|
APOO
|
apolipoprotein O
|
|
chr2_-_130619038
|
0.235
|
NM_207310
|
CCDC74B
|
coiled-coil domain containing 74B
|
|
chr7_+_129693938
|
0.234
|
NM_001869
|
CPA2
|
carboxypeptidase A2 (pancreatic)
|
|
chr3_-_36756355
|
0.233
|
NM_033403
|
DCLK3
|
doublecortin-like kinase 3
|
|
chr12_+_53177806
|
0.232
|
|
NCKAP1L
|
NCK-associated protein 1-like
|
|
chr3_+_108578877
|
0.229
|
NM_032600
|
CCDC54
|
coiled-coil domain containing 54
|
|
chr19_-_63556632
|
0.229
|
NM_130786
|
A1BG
|
alpha-1-B glycoprotein
|
|
chr4_-_75083160
|
0.228
|
NM_002994
|
CXCL5
|
chemokine (C-X-C motif) ligand 5
|
|
chr5_-_43432969
|
0.227
|
|
CCL28
|
chemokine (C-C motif) ligand 28
|
|
chr4_-_122304925
|
0.226
|
NM_024873
|
TNIP3
|
TNFAIP3 interacting protein 3
|
|
chr6_-_2696152
|
0.226
|
NM_001012418
|
MYLK4
|
myosin light chain kinase family, member 4
|
|
chr9_+_35032222
|
0.226
|
NM_001040410
NM_203299
|
C9orf131
|
chromosome 9 open reading frame 131
|