|
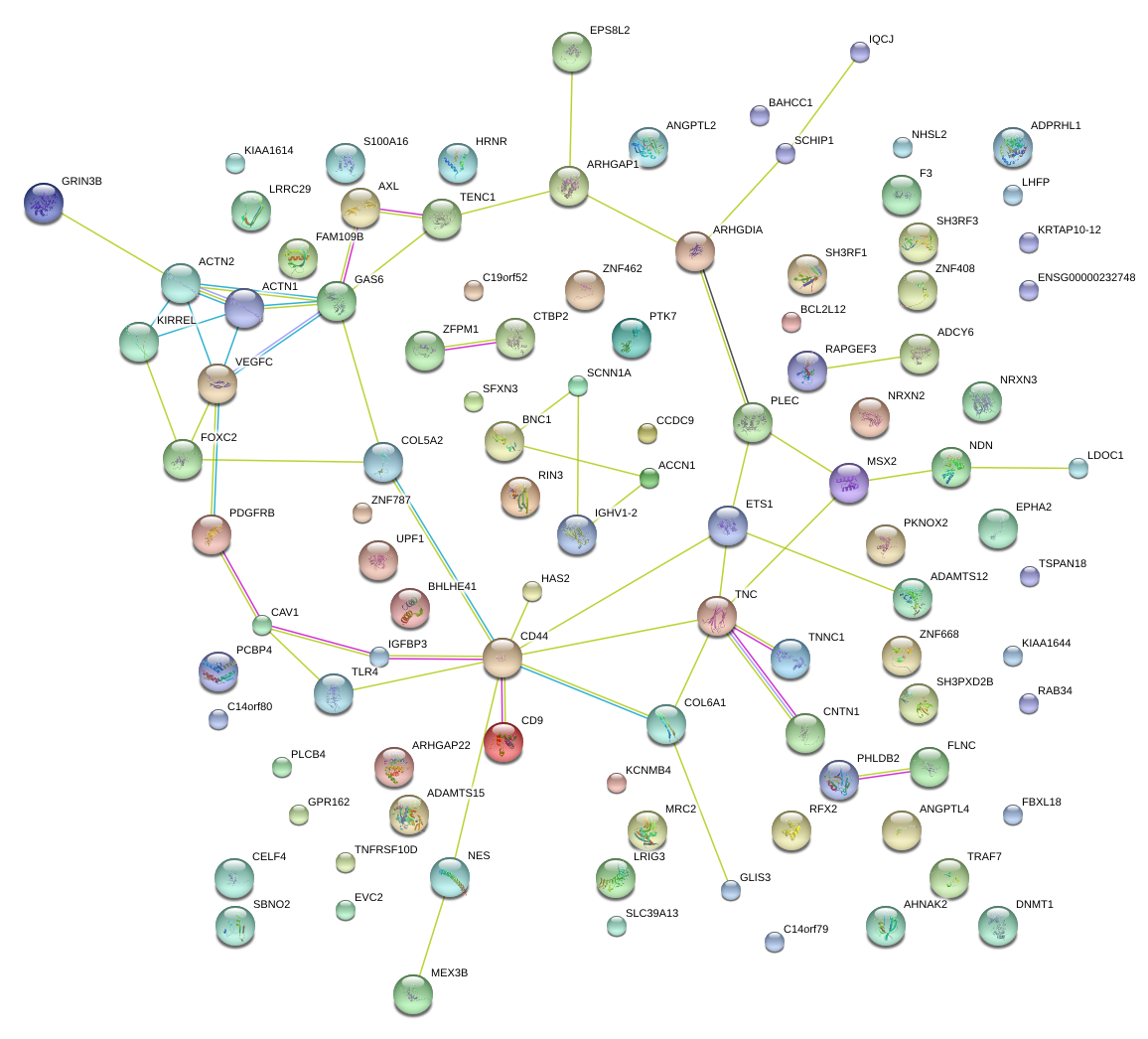
chr12_+_6179736
|
2.199
|
NM_001769
|
CD9
|
CD9 molecule
|
|
chr1_+_156229934
|
2.116
|
|
KIRREL
|
kin of IRRE like (Drosophila)
|
|
chr14_-_104515738
|
1.862
|
NM_138420
|
AHNAK2
|
AHNAK nucleoprotein 2
|
|
chr13_+_113546841
|
1.821
|
NM_000820
|
GAS6
|
growth arrest-specific 6
|
|
chr21_+_44941514
|
1.726
|
NM_198699
|
KRTAP10-12
|
keratin associated protein 10-12
|
|
chr13_+_113546897
|
1.466
|
|
GAS6
|
growth arrest-specific 6
|
|
chr3_+_113061218
|
1.445
|
NM_001134439
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2
|
|
chr3_-_51971915
|
1.444
|
NM_033010
|
PCBP4
|
poly(rC) binding protein 4
|
|
chr1_+_156229686
|
1.393
|
NM_018240
|
KIRREL
|
kin of IRRE like (Drosophila)
|
|
chr3_+_113061330
|
1.326
|
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2
|
|
chr19_-_61324456
|
1.315
|
NM_001002836
|
ZNF787
|
zinc finger protein 787
|
|
chr16_-_65818401
|
1.298
|
NM_001004055
|
LRRC29
|
leucine rich repeat containing 29
|
|
chr8_-_23077392
|
1.259
|
NM_003840
|
TNFRSF10D
|
tumor necrosis factor receptor superfamily, member 10d, decoy with truncated death domain
|
|
chr19_+_10900502
|
1.199
|
|
C19orf52
|
chromosome 19 open reading frame 52
|
|
chr16_-_30992574
|
1.183
|
|
ZNF668
|
zinc finger protein 668
|
|
chr5_+_174084137
|
1.170
|
NM_002449
|
MSX2
|
msh homeobox 2
|
|
chr13_+_113546912
|
1.169
|
|
GAS6
|
growth arrest-specific 6
|
|
chr16_-_30993004
|
1.152
|
NM_024706
|
ZNF668
|
zinc finger protein 668
|
|
chrX_-_140098941
|
1.143
|
|
LDOC1
|
leucine zipper, down-regulated in cancer 1
|
|
chr10_+_102782156
|
1.120
|
|
SFXN3
|
sideroflexin 3
|
|
chr12_+_51727076
|
1.103
|
NM_198316
|
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2)
|
|
chr5_-_33927724
|
1.096
|
NM_030955
|
ADAMTS12
|
ADAM metallopeptidase with thrombospondin type 1 motif, 12
|
|
chr17_-_24068759
|
1.079
|
NM_001144942
NM_031934
|
RAB34
|
RAB34, member RAS oncogene family
|
|
chr9_-_116920232
|
1.051
|
NM_002160
|
TNC
|
tenascin C
|
|
chr22_+_40800193
|
1.037
|
NM_001002034
|
FAM109B
|
family with sequence similarity 109, member B
|
|
chr12_-_47468985
|
1.034
|
NM_020983
|
ADCY6
|
adenylate cyclase 6
|
|
chr22_+_40800213
|
1.032
|
|
FAM109B
|
family with sequence similarity 109, member B
|
|
chr19_+_951295
|
1.030
|
NM_138690
|
GRIN3B
|
glutamate receptor, ionotropic, N-methyl-D-aspartate 3B
|
|
chr15_-_21483503
|
1.028
|
NM_002487
|
NDN
|
necdin homolog (mouse)
|
|
chr4_-_177950658
|
1.024
|
NM_005429
|
VEGFC
|
vascular endothelial growth factor C
|
|
chr12_-_57600564
|
1.021
|
|
LRIG3
|
leucine-rich repeats and immunoglobulin-like domains 3
|
|
chr3_+_113061296
|
1.019
|
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2
|
|
chr11_+_35116961
|
1.001
|
NM_000610
NM_001001389
NM_001001390
NM_001001391
NM_001001392
|
CD44
|
CD44 molecule (Indian blood group)
|
|
chr5_-_149515502
|
0.999
|
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr13_+_113546943
|
0.998
|
|
GAS6
|
growth arrest-specific 6
|
|
chr19_+_52451642
|
0.978
|
|
CCDC9
|
coiled-coil domain containing 9
|
|
chr11_-_64247230
|
0.975
|
NM_015080
NM_138732
|
NRXN2
|
neurexin 2
|
|
chr14_+_92049396
|
0.972
|
NM_024832
|
RIN3
|
Ras and Rab interactor 3
|
|
chr12_+_6808025
|
0.970
|
|
LEPREL2
|
leprecan-like 2
|
|
chr17_+_76988134
|
0.967
|
NM_001080519
|
BAHCC1
|
BAH domain and coiled-coil containing 1
|
|
chr14_+_105028614
|
0.962
|
NM_001134875
NM_001134876
|
C14orf80
|
chromosome 14 open reading frame 80
|
|
chr21_+_46226072
|
0.939
|
NM_001848
|
COL6A1
|
collagen, type VI, alpha 1
|
|
chr15_-_21483457
|
0.932
|
|
NDN
|
necdin homolog (mouse)
|
|
chr19_-_1125263
|
0.920
|
NM_014963
|
SBNO2
|
strawberry notch homolog 2 (Drosophila)
|
|
chr12_-_26169074
|
0.915
|
NM_030762
|
BHLHE41
|
basic helix-loop-helix family, member e41
|
|
chr19_-_1125201
|
0.912
|
|
|
|
|
chr22_-_43040063
|
0.908
|
NM_001099294
|
KIAA1644
|
KIAA1644
|
|
chr1_-_94779727
|
0.895
|
NM_001178096
NM_001993
|
F3
|
coagulation factor III (thromboplastin, tissue factor)
|
|
chr3_+_160964574
|
0.894
|
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr11_+_47386621
|
0.890
|
NM_001128225
NM_152264
|
SLC39A13
|
solute carrier family 39 (zinc transporter), member 13
|
|
chr11_+_124539814
|
0.886
|
|
PKNOX2
|
PBX/knotted 1 homeobox 2
|
|
chr8_-_122722674
|
0.879
|
NM_005328
|
HAS2
|
hyaluronan synthase 2
|
|
chrX_+_71047344
|
0.875
|
NM_001013627
|
NHSL2
|
NHS-like 2
|
|
chr5_-_149515331
|
0.874
|
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chrX_-_140098959
|
0.866
|
NM_012317
|
LDOC1
|
leucine zipper, down-regulated in cancer 1
|
|
chr1_-_16355013
|
0.863
|
NM_004431
|
EPHA2
|
EPH receptor A2
|
|
chr19_-_1125257
|
0.863
|
|
SBNO2
|
strawberry notch homolog 2 (Drosophila)
|
|
chr11_-_46678691
|
0.859
|
NM_004308
|
ARHGAP1
|
Rho GTPase activating protein 1
|
|
chr17_+_58058783
|
0.853
|
|
MRC2
|
mannose receptor, C type 2
|
|
chr11_+_47386707
|
0.853
|
|
SLC39A13
|
solute carrier family 39 (zinc transporter), member 13
|
|
chr11_-_2881750
|
0.848
|
NM_007105
|
SLC22A18AS
|
solute carrier family 22 (organic cation transporter), member 18 antisense
|
|
chr14_-_68515791
|
0.845
|
NM_001102
NM_001130004
NM_001130005
|
ACTN1
|
actinin, alpha 1
|
|
chr11_+_35117226
|
0.841
|
|
CD44
|
CD44 molecule (Indian blood group)
|
|
chr19_+_46417117
|
0.840
|
|
AXL
|
AXL receptor tyrosine kinase
|
|
chr11_-_46678736
|
0.833
|
|
ARHGAP1
|
Rho GTPase activating protein 1
|
|
chr2_-_189752575
|
0.830
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr15_-_80125514
|
0.829
|
|
MEX3B
|
mex-3 homolog B (C. elegans)
|
|
chr13_-_39075304
|
0.826
|
|
LHFP
|
lipoma HMGIC fusion partner
|
|
chr11_+_46678892
|
0.814
|
NM_024741
|
ZNF408
|
zinc finger protein 408
|
|
chr11_+_124539768
|
0.813
|
NM_022062
|
PKNOX2
|
PBX/knotted 1 homeobox 2
|
|
chr10_-_126839546
|
0.809
|
NM_001083914
|
CTBP2
|
C-terminal binding protein 2
|
|
chr7_+_128257698
|
0.807
|
NM_001127487
NM_001458
|
FLNC
|
filamin C, gamma
|
|
chr3_+_113060716
|
0.804
|
NM_001134438
NM_145753
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2
|
|
chr11_+_44742551
|
0.801
|
NM_130783
|
TSPAN18
|
tetraspanin 18
|
|
chr15_-_81743198
|
0.791
|
|
BNC1
|
basonuclin 1
|
|
chr11_+_46679140
|
0.783
|
NM_001184751
|
ZNF408
|
zinc finger protein 408
|
|
chr2_+_109112428
|
0.780
|
NM_001099289
|
SH3RF3
|
SH3 domain containing ring finger 3
|
|
chrX_-_140099014
|
0.771
|
|
LDOC1
|
leucine zipper, down-regulated in cancer 1
|
|
chr9_-_128924706
|
0.769
|
|
ANGPTL2
|
angiopoietin-like 2
|
|
chr4_-_5761194
|
0.759
|
NM_147127
|
EVC2
|
Ellis van Creveld syndrome 2
|
|
chr9_+_108665168
|
0.753
|
NM_021224
|
ZNF462
|
zinc finger protein 462
|
|
chr14_-_68515761
|
0.749
|
|
ACTN1
|
actinin, alpha 1
|
|
chr12_-_57600336
|
0.746
|
NM_153377
|
LRIG3
|
leucine-rich repeats and immunoglobulin-like domains 3
|
|
chr9_-_128924782
|
0.744
|
|
ANGPTL2
|
angiopoietin-like 2
|
|
chr14_-_68515731
|
0.742
|
|
ACTN1
|
actinin, alpha 1
|
|
chr9_-_4290028
|
0.737
|
NM_001042413
|
GLIS3
|
GLIS family zinc finger 3
|
|
chr11_-_127897271
|
0.731
|
NM_001162422
NM_005238
|
ETS1
|
v-ets erythroblastosis virus E26 oncogene homolog 1 (avian)
|
|
chr8_-_145085616
|
0.729
|
NM_201384
|
PLEC
|
plectin
|
|
chr1_-_154913712
|
0.722
|
NM_006617
|
NES
|
nestin
|
|
chr10_-_49483002
|
0.705
|
NM_021226
|
ARHGAP22
|
Rho GTPase activating protein 22
|
|
chr14_-_100420936
|
0.703
|
NM_001134888
|
RTL1
|
retrotransposon-like 1
|
|
chr18_-_33399997
|
0.699
|
NM_001025087
NM_001025088
NM_001025089
NM_020180
|
CELF4
|
CUGBP, Elav-like family member 4
|
|
chr19_+_10900419
|
0.698
|
NM_138358
|
C19orf52
|
chromosome 19 open reading frame 52
|
|
chr14_-_68515742
|
0.697
|
|
ACTN1
|
actinin, alpha 1
|
|
chr6_+_43151922
|
0.697
|
NM_002821
NM_152880
NM_152881
NM_152882
|
PTK7
|
PTK7 protein tyrosine kinase 7
|
|
chr13_-_113151453
|
0.693
|
NM_199162
|
ADPRHL1
|
ADP-ribosylhydrolase like 1
|
|
chr19_+_52451570
|
0.691
|
NM_015603
|
CCDC9
|
coiled-coil domain containing 9
|
|
chr14_-_68515727
|
0.689
|
|
ACTN1
|
actinin, alpha 1
|
|
chr19_-_6061491
|
0.684
|
|
RFX2
|
regulatory factor X, 2 (influences HLA class II expression)
|
|
chr14_-_68515525
|
0.676
|
|
ACTN1
|
actinin, alpha 1
|
|
chr3_+_113060826
|
0.673
|
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2
|
|
chr7_-_45927263
|
0.673
|
NM_000598
NM_001013398
|
IGFBP3
|
insulin-like growth factor binding protein 3
|
|
chr1_+_179148935
|
0.667
|
NM_020950
|
KIAA1614
|
KIAA1614
|
|
chr12_-_46439127
|
0.662
|
NM_001098531
|
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3
|
|
chr7_-_5519831
|
0.661
|
NM_024963
|
FBXL18
|
F-box and leucine-rich repeat protein 18
|
|
chr12_-_6354650
|
0.656
|
NM_001159576
|
SCNN1A
|
sodium channel, nonvoltage-gated 1 alpha
|
|
chr14_+_104523660
|
0.655
|
NM_174891
|
C14orf79
|
chromosome 14 open reading frame 79
|
|
chr12_+_69046241
|
0.655
|
NM_014505
|
KCNMB4
|
potassium large conductance calcium-activated channel, subfamily M, beta member 4
|
|
chr19_-_10166536
|
0.652
|
|
DNMT1
|
DNA (cytosine-5-)-methyltransferase 1
|
|
chr17_-_77422561
|
0.651
|
NM_001185078
NM_004309
|
ARHGDIA
|
Rho GDP dissociation inhibitor (GDI) alpha
|
|
chr11_+_129823667
|
0.650
|
NM_139055
|
ADAMTS15
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15
|
|
chr16_+_85158357
|
0.646
|
NM_005251
|
FOXC2
|
forkhead box C2 (MFH-1, mesenchyme forkhead 1)
|
|
chr11_+_696607
|
0.644
|
|
EPS8L2
|
EPS8-like 2
|
|
chr1_-_151852128
|
0.643
|
NM_080388
|
S100A16
|
S100 calcium binding protein A16
|
|
chr9_+_119506428
|
0.638
|
|
TLR4
|
toll-like receptor 4
|
|
chr16_+_2145765
|
0.629
|
NM_032271
|
TRAF7
|
TNF receptor-associated factor 7
|
|
chr14_-_68515701
|
0.628
|
|
ACTN1
|
actinin, alpha 1
|
|
chr7_+_115952074
|
0.621
|
NM_001753
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chr20_+_9236446
|
0.621
|
NM_000933
|
PLCB4
|
phospholipase C, beta 4
|
|
chr19_+_18803742
|
0.621
|
NM_002911
|
UPF1
|
UPF1 regulator of nonsense transcripts homolog (yeast)
|
|
chr5_-_171813960
|
0.621
|
|
SH3PXD2B
|
SH3 and PX domains 2B
|
|
chr19_+_18803786
|
0.620
|
|
UPF1
|
UPF1 regulator of nonsense transcripts homolog (yeast)
|
|
chr15_-_66511488
|
0.619
|
NM_001004439
|
ITGA11
|
integrin, alpha 11
|
|
chr3_+_156280497
|
0.612
|
|
MME
|
membrane metallo-endopeptidase
|
|
chr17_-_77422400
|
0.612
|
|
|
|
|
chr8_+_104222052
|
0.612
|
NM_001024372
NM_024812
|
BAALC
|
brain and acute leukemia, cytoplasmic
|
|
chr8_+_22078622
|
0.604
|
|
BMP1
|
bone morphogenetic protein 1
|
|
chr10_-_105202139
|
0.602
|
NM_015916
|
CALHM2
|
calcium homeostasis modulator 2
|
|
chr1_-_40009513
|
0.600
|
NM_022120
|
OXCT2
|
3-oxoacid CoA transferase 2
|
|
chr10_-_75352540
|
0.596
|
NM_001001791
|
C10orf55
|
chromosome 10 open reading frame 55
|
|
chr3_+_156280539
|
0.594
|
|
MME
|
membrane metallo-endopeptidase
|
|
chr17_+_40594046
|
0.593
|
NM_144608
|
HEXIM2
|
hexamthylene bis-acetamide inducible 2
|
|
chr16_+_30993223
|
0.591
|
NM_014699
|
ZNF646
|
zinc finger protein 646
|
|
chr22_+_22907443
|
0.586
|
NM_019601
|
SUSD2
|
sushi domain containing 2
|
|
chr8_+_22513043
|
0.585
|
NM_001013842
NM_001198827
NM_173686
|
C8orf58
|
chromosome 8 open reading frame 58
|
|
chr11_+_12355600
|
0.583
|
NM_018222
|
PARVA
|
parvin, alpha
|
|
chr19_-_18913017
|
0.583
|
|
HOMER3
|
homer homolog 3 (Drosophila)
|
|
chr11_-_46678715
|
0.582
|
|
ARHGAP1
|
Rho GTPase activating protein 1
|
|
chr6_+_30018287
|
0.579
|
NM_002116
|
HLA-A
|
major histocompatibility complex, class I, A
|
|
chr3_-_31998241
|
0.579
|
NM_001174060
NM_017784
|
OSBPL10
|
oxysterol binding protein-like 10
|
|
chr16_+_3447957
|
0.577
|
NM_001083600
NM_024845
|
NAT15
|
N-acetyltransferase 15 (GCN5-related, putative)
|
|
chr17_-_72045218
|
0.576
|
NM_134268
|
CYGB
|
cytoglobin
|
|
chr10_-_80875097
|
0.575
|
NM_153367
|
ZCCHC24
|
zinc finger, CCHC domain containing 24
|
|
chr8_-_23317577
|
0.572
|
|
LOXL2
|
lysyl oxidase-like 2
|
|
chr11_+_1925075
|
0.571
|
NM_021134
|
MRPL23
|
mitochondrial ribosomal protein L23
|
|
chr17_+_76003574
|
0.570
|
|
FLJ35220
|
hypothetical protein FLJ35220
|
|
chr18_+_19973445
|
0.570
|
NM_138644
|
CABYR
|
calcium binding tyrosine-(Y)-phosphorylation regulated
|
|
chr9_-_4289915
|
0.569
|
|
GLIS3
|
GLIS family zinc finger 3
|
|
chr19_-_18749728
|
0.568
|
|
|
|
|
chr16_+_30993252
|
0.568
|
|
ZNF646
|
zinc finger protein 646
|
|
chr19_-_18913011
|
0.566
|
|
HOMER3
|
homer homolog 3 (Drosophila)
|
|
chr1_+_182622751
|
0.565
|
NM_030806
|
C1orf21
|
chromosome 1 open reading frame 21
|
|
chr10_-_49534304
|
0.565
|
|
ARHGAP22
|
Rho GTPase activating protein 22
|
|
chr1_-_120413798
|
0.562
|
NM_024408
|
NOTCH2
|
notch 2
|
|
chr16_-_65818221
|
0.560
|
NM_012163
|
LRRC29
|
leucine rich repeat containing 29
|
|
chr3_-_51976428
|
0.559
|
NM_020418
NM_001174100
NM_033008
|
PCBP4
|
poly(rC) binding protein 4
|
|
chr22_+_36286350
|
0.558
|
NM_152243
|
CDC42EP1
|
CDC42 effector protein (Rho GTPase binding) 1
|
|
chr2_-_40532651
|
0.558
|
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1
|
|
chr3_+_156280129
|
0.555
|
NM_000902
|
MME
|
membrane metallo-endopeptidase
|
|
chr5_+_140747586
|
0.555
|
NM_003736
NM_032098
|
PCDHGB4
|
protocadherin gamma subfamily B, 4
|
|
chr22_-_27405708
|
0.555
|
NM_001145418
|
TTC28
|
tetratricopeptide repeat domain 28
|
|
chr6_-_72187168
|
0.553
|
|
C6orf155
|
chromosome 6 open reading frame 155
|
|
chr8_-_119192985
|
0.550
|
NM_000127
|
EXT1
|
exostosin 1
|
|
chr14_-_88090641
|
0.549
|
NM_007039
|
PTPN21
|
protein tyrosine phosphatase, non-receptor type 21
|
|
chr8_-_23317571
|
0.549
|
|
LOXL2
|
lysyl oxidase-like 2
|
|
chr19_+_12966970
|
0.549
|
NM_002501
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr19_-_18913032
|
0.547
|
NM_004838
|
HOMER3
|
homer homolog 3 (Drosophila)
|
|
chr6_+_46763570
|
0.547
|
NM_001010870
NM_001168359
|
TDRD6
|
tudor domain containing 6
|
|
chr15_-_84139021
|
0.546
|
|
KLHL25
|
kelch-like 25 (Drosophila)
|
|
chr1_+_182622850
|
0.545
|
|
C1orf21
|
chromosome 1 open reading frame 21
|
|
chr9_+_129509088
|
0.545
|
NM_001012502
|
C9orf117
|
chromosome 9 open reading frame 117
|
|
chr1_-_56817717
|
0.543
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr16_+_639278
|
0.543
|
NM_145294
|
WDR90
|
WD repeat domain 90
|
|
chr10_+_114699953
|
0.542
|
NM_001146274
NM_001146283
NM_001146284
NM_001146285
NM_001146286
NM_001198525
NM_001198526
NM_001198527
NM_001198528
NM_001198529
NM_001198530
NM_001198531
NM_030756
|
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box)
|
|
chr1_-_93919789
|
0.541
|
NM_003567
|
BCAR3
|
breast cancer anti-estrogen resistance 3
|
|
chr18_-_20231709
|
0.540
|
|
OSBPL1A
|
oxysterol binding protein-like 1A
|
|
chr5_+_119827797
|
0.540
|
|
PRR16
|
proline rich 16
|
|
chr1_-_1233109
|
0.540
|
NM_030649
|
ACAP3
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3
|
|
chr8_+_22492198
|
0.539
|
NM_021630
|
PDLIM2
|
PDZ and LIM domain 2 (mystique)
|
|
chr1_-_199613427
|
0.538
|
NM_000364
NM_001001430
NM_001001431
NM_001001432
|
TNNT2
|
troponin T type 2 (cardiac)
|
|
chr2_+_28469275
|
0.537
|
NM_005253
|
FOSL2
|
FOS-like antigen 2
|
|
chr1_+_182622809
|
0.536
|
|
C1orf21
|
chromosome 1 open reading frame 21
|
|
chr1_+_23990228
|
0.533
|
NM_007260
|
LYPLA2
|
lysophospholipase II
|
|
chr6_-_112682459
|
0.532
|
NM_001105206
NM_001105207
NM_001105208
NM_001105209
NM_002290
|
LAMA4
|
laminin, alpha 4
|
|
chr7_+_115952279
|
0.527
|
NM_001172895
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chr8_-_23317536
|
0.527
|
|
LOXL2
|
lysyl oxidase-like 2
|
|
chr22_+_22907417
|
0.526
|
|
SUSD2
|
sushi domain containing 2
|
|
chr18_-_20231783
|
0.521
|
NM_080597
|
OSBPL1A
|
oxysterol binding protein-like 1A
|
|
chr19_+_4255590
|
0.520
|
NM_024333
|
FSD1
|
fibronectin type III and SPRY domain containing 1
|
|
chr8_-_26427304
|
0.519
|
NM_007257
|
PNMA2
|
paraneoplastic antigen MA2
|
|
chr10_+_134060681
|
0.518
|
NM_001098637
NM_138499
|
PWWP2B
|
PWWP domain containing 2B
|
|
chr19_-_8485026
|
0.516
|
NM_001146175
NM_032370
|
ZNF414
|
zinc finger protein 414
|
|
chrX_+_152684150
|
0.516
|
|
PLXNB3
|
plexin B3
|
|
chr15_-_80125403
|
0.516
|
NM_032246
|
MEX3B
|
mex-3 homolog B (C. elegans)
|
|
chr1_+_112852892
|
0.515
|
NM_024494
|
WNT2B
|
wingless-type MMTV integration site family, member 2B
|
|
chr14_-_104333060
|
0.514
|
|
AKT1
|
v-akt murine thymoma viral oncogene homolog 1
|
|
chr1_+_53300410
|
0.513
|
NM_153703
|
PODN
|
podocan
|
|
chr10_+_102782151
|
0.512
|
|
SFXN3
|
sideroflexin 3
|
|
chr17_-_68819713
|
0.511
|
NM_012121
|
CDC42EP4
|
CDC42 effector protein (Rho GTPase binding) 4
|
|
chr2_-_218843109
|
0.510
|
NM_001087
|
AAMP
|
angio-associated, migratory cell protein
|