|
chr22_+_35586950
|
2.554
|
NM_000631
NM_013416
|
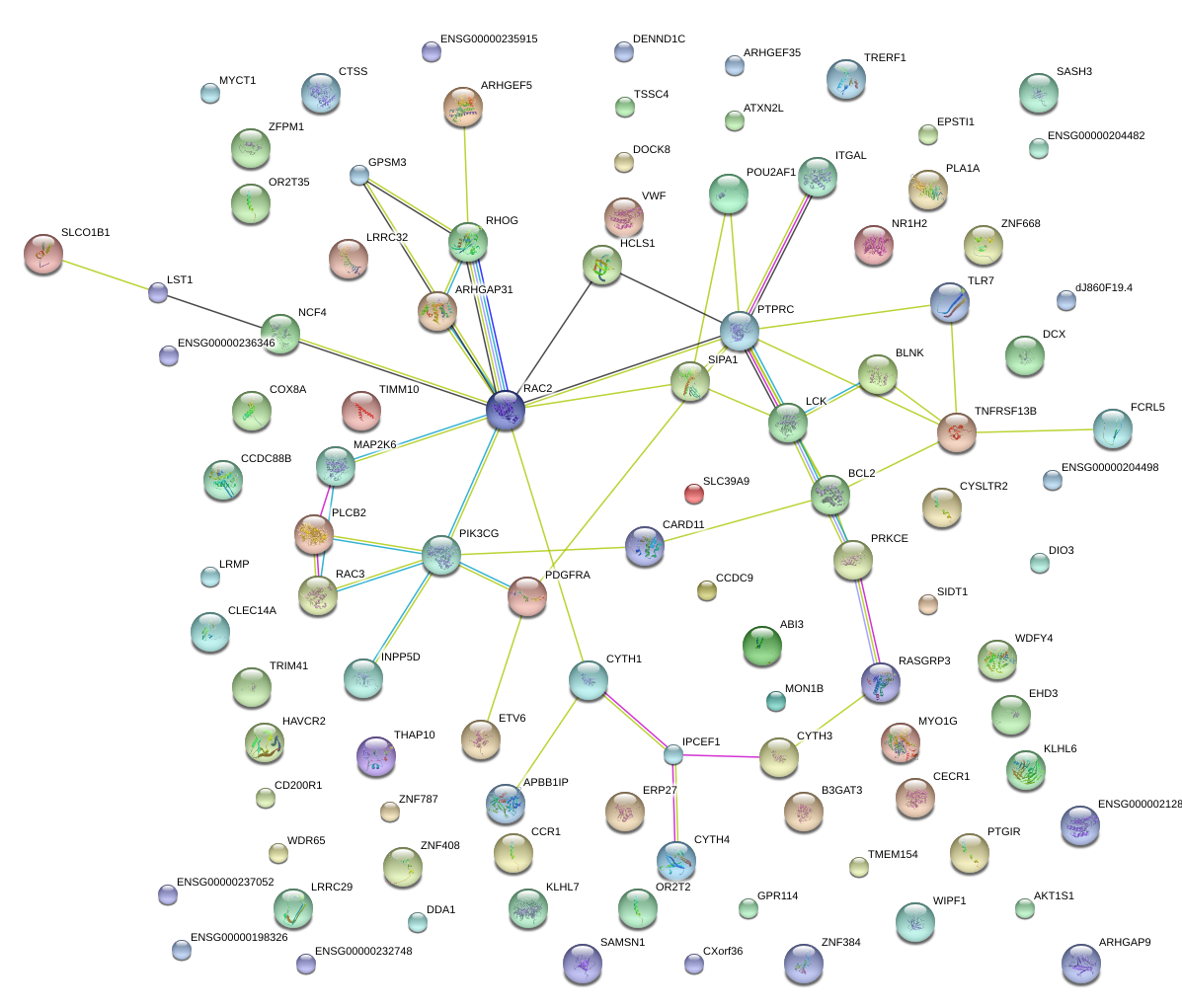
NCF4
|
neutrophil cytosolic factor 4, 40kDa
|
|
chr10_+_49562866
|
2.271
|
|
WDFY4
|
WDFY family member 4
|
|
chr7_+_106292975
|
2.235
|
|
PIK3CG
|
phosphoinositide-3-kinase, catalytic, gamma polypeptide
|
|
chr7_+_106292932
|
2.122
|
|
PIK3CG
|
phosphoinositide-3-kinase, catalytic, gamma polypeptide
|
|
chr3_+_114733907
|
2.088
|
NM_017699
|
SIDT1
|
SID1 transmembrane family, member 1
|
|
chr10_-_98021144
|
1.980
|
NM_001114094
NM_013314
|
BLNK
|
B-cell linker
|
|
chr3_-_122862391
|
1.936
|
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr3_-_122862447
|
1.897
|
NM_005335
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr12_-_6668880
|
1.814
|
NM_001039917
NM_001039919
NM_001039920
NM_001135734
|
ZNF384
|
zinc finger protein 384
|
|
chr3_+_120495906
|
1.813
|
NM_020754
|
ARHGAP31
|
Rho GTPase activating protein 31
|
|
chr3_-_46224791
|
1.803
|
NM_001295
|
CCR1
|
chemokine (C-C motif) receptor 1
|
|
chr7_+_106293112
|
1.744
|
NM_002649
|
PIK3CG
|
phosphoinositide-3-kinase, catalytic, gamma polypeptide
|
|
chr12_-_6668680
|
1.697
|
|
ZNF384
|
zinc finger protein 384
|
|
chr11_+_63498590
|
1.603
|
NM_004074
|
COX8A
|
cytochrome c oxidase subunit VIIIA (ubiquitous)
|
|
chr12_-_6668787
|
1.584
|
NM_001039918
NM_133476
|
ZNF384
|
zinc finger protein 384
|
|
chr6_-_42527760
|
1.583
|
NM_033502
|
TRERF1
|
transcriptional regulating factor 1
|
|
chr18_-_59137488
|
1.566
|
NM_000633
NM_000657
|
BCL2
|
B-cell CLL/lymphoma 2
|
|
chr19_-_61324456
|
1.555
|
NM_001002836
|
ZNF787
|
zinc finger protein 787
|
|
chr17_-_16816113
|
1.517
|
NM_012452
|
TNFRSF13B
|
tumor necrosis factor receptor superfamily, member 13B
|
|
chr1_+_32512298
|
1.511
|
NM_001042771
|
LCK
|
lymphocyte-specific protein tyrosine kinase
|
|
chr3_-_184756101
|
1.502
|
NM_130446
|
KLHL6
|
kelch-like 6 (Drosophila)
|
|
chr19_-_55071645
|
1.465
|
|
AKT1S1
|
AKT1 substrate 1 (proline-rich)
|
|
chr3_+_120799411
|
1.429
|
NM_015900
|
PLA1A
|
phospholipase A1 member A
|
|
chr11_+_2378559
|
1.418
|
|
TSSC4
|
tumor suppressing subtransferable candidate 4
|
|
chr22_+_36008465
|
1.400
|
|
CYTH4
|
cytohesin 4
|
|
chr19_-_51820133
|
1.390
|
NM_000960
|
PTGIR
|
prostaglandin I2 (prostacyclin) receptor (IP)
|
|
chr2_-_175207521
|
1.384
|
NM_003387
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr16_-_30993004
|
1.374
|
NM_024706
|
ZNF668
|
zinc finger protein 668
|
|
chr6_-_32268267
|
1.371
|
|
GPSM3
|
G-protein signaling modulator 3
|
|
chr19_+_17281374
|
1.362
|
|
DDA1
|
DET1 and DDB1 associated 1
|
|
chr10_+_49563520
|
1.326
|
NM_020945
|
WDFY4
|
WDFY family member 4
|
|
chr17_+_44642587
|
1.320
|
NM_001135186
NM_016428
|
ABI3
|
ABI family, member 3
|
|
chr17_+_64922420
|
1.315
|
NM_002758
|
MAP2K6
|
mitogen-activated protein kinase kinase 6
|
|
chrX_+_128741562
|
1.313
|
NM_018990
|
SASH3
|
SAM and SH3 domain containing 3
|
|
chr1_-_149004879
|
1.306
|
|
CTSS
|
cathepsin S
|
|
chr14_+_101097440
|
1.301
|
NM_001362
|
DIO3
|
deiodinase, iodothyronine, type III
|
|
chr22_+_25398690
|
1.294
|
|
LOC388889
|
hypothetical LOC388889
|
|
chr1_+_196874867
|
1.285
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr11_-_76059435
|
1.284
|
NM_001128922
|
LRRC32
|
leucine rich repeat containing 32
|
|
chr11_-_62146144
|
1.270
|
|
B3GAT3
|
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I)
|
|
chr16_-_65818401
|
1.265
|
NM_001004055
|
LRRC29
|
leucine rich repeat containing 29
|
|
chr12_-_56157898
|
1.263
|
NM_001080156
|
ARHGAP9
|
Rho GTPase activating protein 9
|
|
chr2_+_33514896
|
1.231
|
NM_170672
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated)
|
|
chr6_-_32268615
|
1.222
|
|
GPSM3
|
G-protein signaling modulator 3
|
|
chr4_+_54790020
|
1.221
|
NM_006206
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr4_-_153820535
|
1.216
|
NM_152680
|
TMEM154
|
transmembrane protein 154
|
|
chr1_+_32489426
|
1.210
|
NM_005356
|
LCK
|
lymphocyte-specific protein tyrosine kinase
|
|
chr6_-_154719560
|
1.209
|
NM_001130700
NM_015553
|
IPCEF1
|
interaction protein for cytohesin exchange factors 1
|
|
chr1_+_43410587
|
1.205
|
NM_001167965
NM_152498
NM_001167966
|
WDR65
|
WD repeat domain 65
|
|
chr16_-_30992574
|
1.194
|
|
ZNF668
|
zinc finger protein 668
|
|
chr19_+_55571493
|
1.189
|
NM_007121
|
NR1H2
|
nuclear receptor subfamily 1, group H, member 2
|
|
chr16_+_56134037
|
1.183
|
|
GPR114
|
G protein-coupled receptor 114
|
|
chr16_+_30391460
|
1.176
|
NM_001114380
NM_002209
|
ITGAL
|
integrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide)
|
|
chr15_-_68971665
|
1.165
|
NM_020147
|
THAP10
|
THAP domain containing 10
|
|
chrX_+_128741578
|
1.158
|
|
SASH3
|
SAM and SH3 domain containing 3
|
|
chr11_+_46679140
|
1.154
|
NM_001184751
|
ZNF408
|
zinc finger protein 408
|
|
chr16_+_30391550
|
1.150
|
|
ITGAL
|
integrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide)
|
|
chr3_-_114176401
|
1.148
|
NM_138806
NM_138939
NM_138940
NM_170780
|
CD200R1
|
CD200 receptor 1
|
|
chr22_+_36008511
|
1.144
|
|
CYTH4
|
cytohesin 4
|
|
chrX_+_12795111
|
1.139
|
NM_016562
|
TLR7
|
toll-like receptor 7
|
|
chr7_-_3049872
|
1.136
|
NM_032415
|
CARD11
|
caspase recruitment domain family, member 11
|
|
chr19_-_6432765
|
1.130
|
NM_024898
|
DENND1C
|
DENN/MADD domain containing 1C
|
|
chr22_+_36008369
|
1.125
|
NM_013385
|
CYTH4
|
cytohesin 4
|
|
chr1_-_246869181
|
1.092
|
NM_001001827
|
OR2T35
|
olfactory receptor, family 2, subfamily T, member 35
|
|
chr21_-_14840506
|
1.089
|
NM_022136
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1
|
|
chr10_+_26767657
|
1.089
|
|
APBB1IP
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein
|
|
chr12_-_56159631
|
1.085
|
NM_001080157
|
ARHGAP9
|
Rho GTPase activating protein 9
|
|
chr2_+_233633279
|
1.082
|
NM_001017915
NM_005541
|
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa
|
|
chr5_+_180582880
|
1.073
|
|
TRIM41
|
tripartite motif containing 41
|
|
chr22_-_35970230
|
1.072
|
NM_002872
|
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2)
|
|
chr12_-_14982708
|
1.067
|
NM_152321
|
ERP27
|
endoplasmic reticulum protein 27
|
|
chr15_-_38387410
|
1.064
|
NM_004573
|
PLCB2
|
phospholipase C, beta 2
|
|
chr13_-_42464301
|
1.062
|
NM_001002264
NM_033255
|
EPSTI1
|
epithelial stromal interaction 1 (breast)
|
|
chr10_+_26767698
|
1.053
|
|
APBB1IP
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein
|
|
chr6_+_31661934
|
1.035
|
NM_205838
NM_205839
|
LST1
|
leukocyte specific transcript 1
|
|
chr13_+_48178620
|
1.035
|
NM_020377
|
CYSLTR2
|
cysteinyl leukotriene receptor 2
|
|
chr15_-_38387164
|
1.033
|
|
PLCB2
|
phospholipase C, beta 2
|
|
chr1_-_155788775
|
1.030
|
NM_001195388
NM_031281
|
FCRL5
|
Fc receptor-like 5
|
|
chr2_+_45732416
|
1.025
|
NM_005400
|
PRKCE
|
protein kinase C, epsilon
|
|
chr16_-_85099941
|
1.022
|
|
LOC400550
|
hypothetical LOC400550
|
|
chr12_+_11694172
|
1.020
|
|
ETV6
|
ets variant 6
|
|
chr16_+_75782870
|
1.015
|
|
MON1B
|
MON1 homolog B (yeast)
|
|
chr11_+_65164167
|
1.009
|
NM_153253
|
SIPA1
|
signal-induced proliferation-associated 1
|
|
chr12_+_25096472
|
1.007
|
NM_006152
|
LRMP
|
lymphoid-restricted membrane protein
|
|
chr15_-_38387317
|
1.003
|
|
PLCB2
|
phospholipase C, beta 2
|
|
chrX_-_44945025
|
1.000
|
NM_024689
NM_176819
|
CXorf36
|
chromosome X open reading frame 36
|
|
chr14_-_37795290
|
0.986
|
NM_175060
|
CLEC14A
|
C-type lectin domain family 14, member A
|
|
chr11_-_62145952
|
0.981
|
NM_012200
|
B3GAT3
|
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I)
|
|
chr7_-_44985202
|
0.980
|
NM_033054
|
MYO1G
|
myosin IG
|
|
chr11_+_63864263
|
0.977
|
NM_032251
|
CCDC88B
|
coiled-coil domain containing 88B
|
|
chr9_+_263025
|
0.972
|
NM_001190458
NM_001193536
|
DOCK8
|
dedicator of cytokinesis 8
|
|
chr2_+_233633348
|
0.968
|
|
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa
|
|
chr22_-_16080265
|
0.964
|
|
CECR1
|
cat eye syndrome chromosome region, candidate 1
|
|
chr5_-_156468614
|
0.961
|
NM_032782
|
HAVCR2
|
hepatitis A virus cellular receptor 2
|
|
chr3_+_120496180
|
0.952
|
|
ARHGAP31
|
Rho GTPase activating protein 31
|
|
chr6_+_153060722
|
0.948
|
NM_025107
|
MYCT1
|
myc target 1
|
|
chr19_+_52451642
|
0.947
|
|
CCDC9
|
coiled-coil domain containing 9
|
|
chrX_-_110542040
|
0.946
|
NM_178151
NM_001195553
NM_178152
NM_178153
|
DCX
|
doublecortin
|
|
chr2_+_31310485
|
0.944
|
|
EHD3
|
EH-domain containing 3
|
|
chr14_+_68934855
|
0.943
|
|
SLC39A9
|
solute carrier family 39 (zinc transporter), member 9
|
|
chr20_+_2743613
|
0.940
|
NM_080739
|
C20orf141
|
chromosome 20 open reading frame 141
|
|
chr12_+_25096755
|
0.939
|
|
LRMP
|
lymphoid-restricted membrane protein
|
|
chr1_+_196874839
|
0.934
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr19_+_52451570
|
0.933
|
NM_015603
|
CCDC9
|
coiled-coil domain containing 9
|
|
chr11_-_110755112
|
0.930
|
|
POU2AF1
|
POU class 2 associating factor 1
|
|
chr12_-_6103945
|
0.926
|
NM_000552
|
VWF
|
von Willebrand factor
|
|
chr7_-_143523582
|
0.914
|
NM_001003702
|
ARHGEF35
|
Rho guanine nucleotide exchange factor (GEF) 35
|
|
chr1_+_196874759
|
0.911
|
NM_002838
NM_080921
NM_080923
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr11_-_57054756
|
0.911
|
NM_012456
|
TIMM10
|
translocase of inner mitochondrial membrane 10 homolog (yeast)
|
|
chr4_+_54790190
|
0.905
|
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr16_-_49272617
|
0.903
|
|
SNX20
|
sorting nexin 20
|
|
chr1_-_158815850
|
0.900
|
|
CD84
|
CD84 molecule
|
|
chr16_-_29664765
|
0.896
|
NM_175900
|
C16orf54
|
chromosome 16 open reading frame 54
|
|
chr11_-_64641700
|
0.894
|
NM_014205
|
ZNHIT2
|
zinc finger, HIT-type containing 2
|
|
chr11_-_46678715
|
0.890
|
|
ARHGAP1
|
Rho GTPase activating protein 1
|
|
chr16_+_56134077
|
0.886
|
NM_153837
|
GPR114
|
G protein-coupled receptor 114
|
|
chr19_+_17281352
|
0.886
|
|
DDA1
|
DET1 and DDB1 associated 1
|
|
chr19_-_59019240
|
0.885
|
NM_144687
|
NLRP12
|
NLR family, pyrin domain containing 12
|
|
chr1_+_111544469
|
0.885
|
|
|
|
|
chr13_-_27967215
|
0.876
|
NM_001159920
NM_001160030
NM_001160031
NM_002019
|
FLT1
|
fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor)
|
|
chr13_-_45654327
|
0.875
|
NM_002298
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr5_-_169657312
|
0.874
|
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr7_-_44985175
|
0.867
|
|
MYO1G
|
myosin IG
|
|
chr19_-_56567602
|
0.866
|
NM_005601
|
NKG7
|
natural killer cell group 7 sequence
|
|
chr13_-_45654296
|
0.864
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr19_-_44518465
|
0.859
|
|
GMFG
|
glia maturation factor, gamma
|
|
chrX_-_102234606
|
0.858
|
NM_022052
|
NXF3
|
nuclear RNA export factor 3
|
|
chr19_-_17349096
|
0.855
|
NM_031310
|
PLVAP
|
plasmalemma vesicle associated protein
|
|
chr17_-_4783943
|
0.848
|
NM_001165417
NM_001165418
NM_003562
|
SLC25A11
|
solute carrier family 25 (mitochondrial carrier; oxoglutarate carrier), member 11
|
|
chr9_-_90983501
|
0.846
|
NM_016848
|
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3
|
|
chr17_-_31441390
|
0.846
|
NM_002983
|
CCL3
|
chemokine (C-C motif) ligand 3
|
|
chr1_+_202000906
|
0.832
|
NM_001136190
NM_017773
|
LAX1
|
lymphocyte transmembrane adaptor 1
|
|
chr7_-_44985084
|
0.824
|
|
MYO1G
|
myosin IG
|
|
chr3_-_150533948
|
0.824
|
NM_001184723
NM_138786
|
TM4SF18
|
transmembrane 4 L six family member 18
|
|
chr1_-_158815895
|
0.823
|
NM_001184879
NM_001184881
NM_001184882
NM_003874
|
CD84
|
CD84 molecule
|
|
chr19_+_10820105
|
0.818
|
NM_001136482
|
C19orf38
|
chromosome 19 open reading frame 38
|
|
chr1_-_181826292
|
0.818
|
NM_000433
NM_001190789
NM_001190794
|
NCF2
|
neutrophil cytosolic factor 2
|
|
chr7_+_143683365
|
0.812
|
NM_005435
|
ARHGEF5
|
Rho guanine nucleotide exchange factor (GEF) 5
|
|
chr18_+_21967813
|
0.806
|
NM_001025096
NM_001025097
NM_144662
|
PSMA8
|
proteasome (prosome, macropain) subunit, alpha type, 8
|
|
chr20_+_2224612
|
0.805
|
NM_003245
|
TGM3
|
transglutaminase 3 (E polypeptide, protein-glutamine-gamma-glutamyltransferase)
|
|
chr19_-_47328399
|
0.802
|
NM_002698
|
POU2F2
|
POU class 2 homeobox 2
|
|
chr5_-_66528330
|
0.801
|
NM_005582
|
CD180
|
CD180 molecule
|
|
chr12_+_11693909
|
0.800
|
|
ETV6
|
ets variant 6
|
|
chr1_-_181826086
|
0.796
|
|
NCF2
|
neutrophil cytosolic factor 2
|
|
chr16_+_2145810
|
0.794
|
|
TRAF7
|
TNF receptor-associated factor 7
|
|
chr12_+_11693993
|
0.791
|
NM_001987
|
ETV6
|
ets variant 6
|
|
chr9_+_94766379
|
0.788
|
|
FGD3
|
FYVE, RhoGEF and PH domain containing 3
|
|
chr17_+_6858779
|
0.787
|
NM_001142798
NM_001142799
NM_174893
|
C17orf49
|
chromosome 17 open reading frame 49
|
|
chr11_-_110755366
|
0.785
|
NM_006235
|
POU2AF1
|
POU class 2 associating factor 1
|
|
chr1_-_149004852
|
0.781
|
|
CTSS
|
cathepsin S
|
|
chr14_+_62740878
|
0.779
|
NM_020663
|
RHOJ
|
ras homolog gene family, member J
|
|
chr4_-_10295412
|
0.777
|
NM_052964
|
CLNK
|
cytokine-dependent hematopoietic cell linker
|
|
chr6_+_10663934
|
0.775
|
NM_001491
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group)
|
|
chr1_+_159943385
|
0.772
|
NM_001184866
NM_001184867
NM_001184870
NM_001184871
NM_001184872
NM_001184873
NM_032738
|
FCRLA
|
Fc receptor-like A
|
|
chr3_-_166397142
|
0.772
|
NM_014926
|
SLITRK3
|
SLIT and NTRK-like family, member 3
|
|
chr19_-_15436333
|
0.771
|
NM_022904
|
RASAL3
|
RAS protein activator like 3
|
|
chr12_-_56422120
|
0.770
|
NM_014770
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2
|
|
chr9_-_116190038
|
0.764
|
|
AKNA
|
AT-hook transcription factor
|
|
chr11_+_63362922
|
0.764
|
NM_001039469
NM_001163296
NM_001163297
NM_004954
|
MARK2
|
MAP/microtubule affinity-regulating kinase 2
|
|
chr1_-_155834357
|
0.762
|
NM_031282
|
FCRL4
|
Fc receptor-like 4
|
|
chr16_+_75782556
|
0.759
|
|
MON1B
|
MON1 homolog B (yeast)
|
|
chr5_+_180582905
|
0.757
|
NM_033549
NM_201627
|
TRIM41
|
tripartite motif containing 41
|
|
chr16_+_2145765
|
0.757
|
NM_032271
|
TRAF7
|
TNF receptor-associated factor 7
|
|
chr20_+_44180355
|
0.757
|
|
CD40
|
CD40 molecule, TNF receptor superfamily member 5
|
|
chr1_+_43539239
|
0.756
|
NM_005424
|
TIE1
|
tyrosine kinase with immunoglobulin-like and EGF-like domains 1
|
|
chr5_-_54317102
|
0.755
|
NM_001135604
NM_007036
|
ESM1
|
endothelial cell-specific molecule 1
|
|
chr21_-_38792173
|
0.754
|
NM_001136155
NM_182918
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr17_-_77422526
|
0.749
|
|
ARHGDIA
|
Rho GDP dissociation inhibitor (GDI) alpha
|
|
chr5_-_38881636
|
0.748
|
|
|
|
|
chr2_-_215605054
|
0.747
|
NM_015657
|
ABCA12
|
ATP-binding cassette, sub-family A (ABC1), member 12
|
|
chr19_+_14412087
|
0.743
|
|
PKN1
|
protein kinase N1
|
|
chr1_-_159306210
|
0.741
|
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr4_+_100957003
|
0.740
|
NM_014395
|
DAPP1
|
dual adaptor of phosphotyrosine and 3-phosphoinositides
|
|
chrX_-_128616594
|
0.738
|
NM_017413
|
APLN
|
apelin
|
|
chr14_+_68935172
|
0.734
|
|
SLC39A9
|
solute carrier family 39 (zinc transporter), member 9
|
|
chr7_+_149842850
|
0.731
|
NM_153236
|
GIMAP7
|
GTPase, IMAP family member 7
|
|
chr7_-_143622188
|
0.731
|
|
ARHGEF5
|
Rho guanine nucleotide exchange factor (GEF) 5
|
|
chr11_+_46678892
|
0.727
|
NM_024741
|
ZNF408
|
zinc finger protein 408
|
|
chr4_+_2783764
|
0.726
|
|
SH3BP2
|
SH3-domain binding protein 2
|
|
chr13_-_45654292
|
0.723
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr16_+_75782595
|
0.722
|
|
MON1B
|
MON1 homolog B (yeast)
|
|
chr11_+_123398929
|
0.718
|
NM_001001953
|
OR10G9
|
olfactory receptor, family 10, subfamily G, member 9
|
|
chr5_+_180583315
|
0.718
|
|
TRIM41
|
tripartite motif containing 41
|
|
chr16_-_49272741
|
0.716
|
NM_001144972
NM_153337
NM_182854
|
SNX20
|
sorting nexin 20
|
|
chr2_-_73373956
|
0.714
|
NM_001965
|
EGR4
|
early growth response 4
|
|
chr17_-_18890963
|
0.710
|
NM_006613
|
GRAP
|
GRB2-related adaptor protein
|
|
chr17_-_59451662
|
0.710
|
NM_001099786
NM_001099787
NM_001099788
NM_001099789
|
ICAM2
|
intercellular adhesion molecule 2
|
|
chr17_+_4283967
|
0.704
|
NM_182538
|
SPNS3
|
spinster homolog 3 (Drosophila)
|
|
chr12_+_106692291
|
0.700
|
NM_203436
|
ASCL4
|
achaete-scute complex homolog 4 (Drosophila)
|
|
chr16_+_29582069
|
0.699
|
NM_003123
|
SPN
|
sialophorin
|
|
chr1_+_179719338
|
0.699
|
NM_000721
|
CACNA1E
|
calcium channel, voltage-dependent, R type, alpha 1E subunit
|
|
chr3_+_123817151
|
0.697
|
NM_152615
|
PARP15
|
poly (ADP-ribose) polymerase family, member 15
|
|
chr2_-_213724577
|
0.694
|
NM_001079526
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr11_+_59979857
|
0.691
|
NM_021950
NM_152866
|
MS4A1
|
membrane-spanning 4-domains, subfamily A, member 1
|
|
chr14_-_104602784
|
0.689
|
NM_013345
|
GPR132
|
G protein-coupled receptor 132
|
|
chr5_-_169657192
|
0.688
|
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr11_-_74818685
|
0.688
|
|
KLHL35
|
kelch-like 35 (Drosophila)
|
|
chr19_-_10166536
|
0.686
|
|
DNMT1
|
DNA (cytosine-5-)-methyltransferase 1
|
|
chr12_+_47162552
|
0.685
|
NM_152319
|
C12orf54
|
chromosome 12 open reading frame 54
|
|
chr14_-_104602809
|
0.684
|
|
GPR132
|
G protein-coupled receptor 132
|