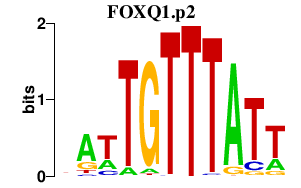
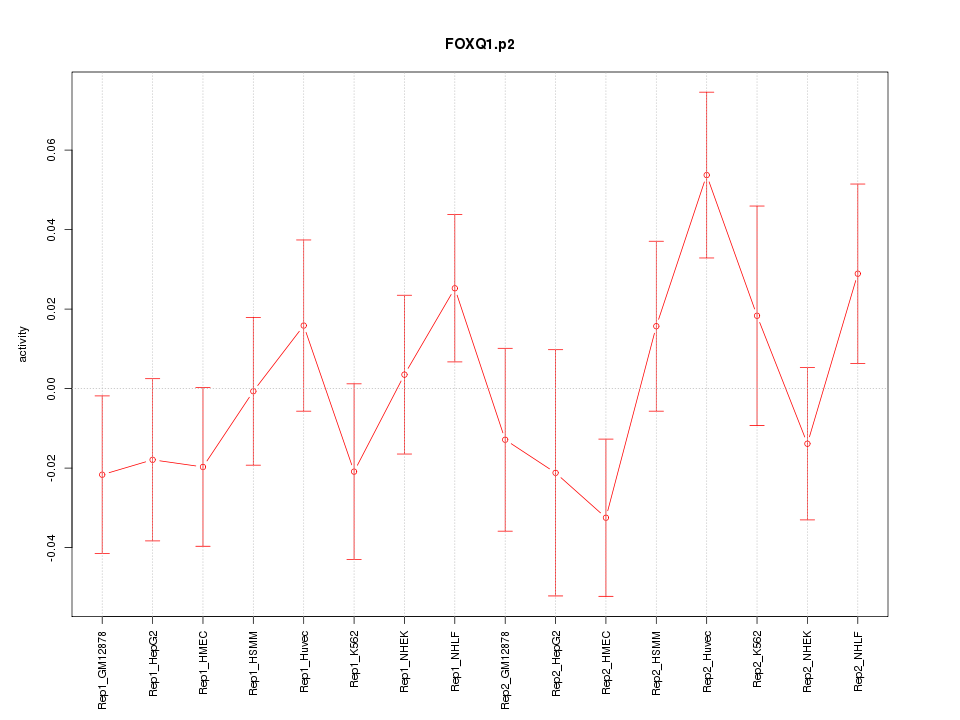
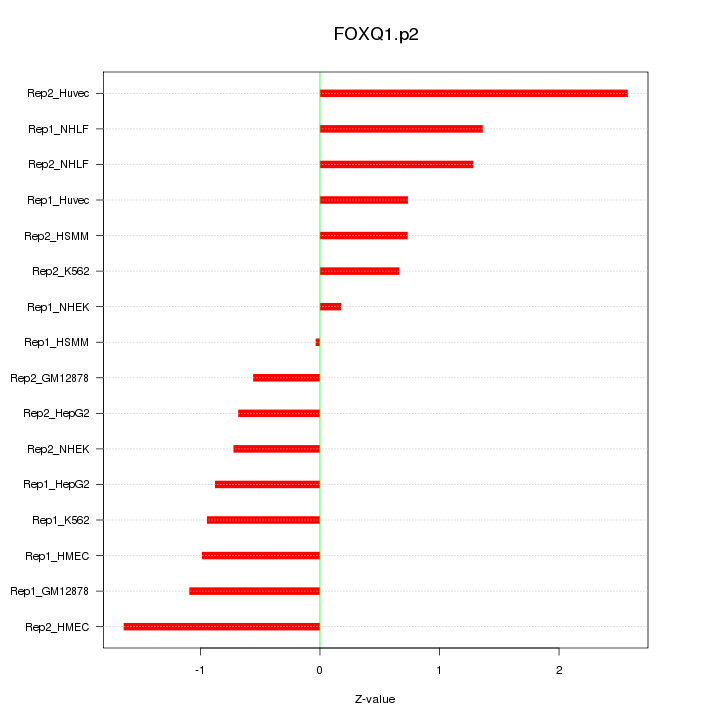
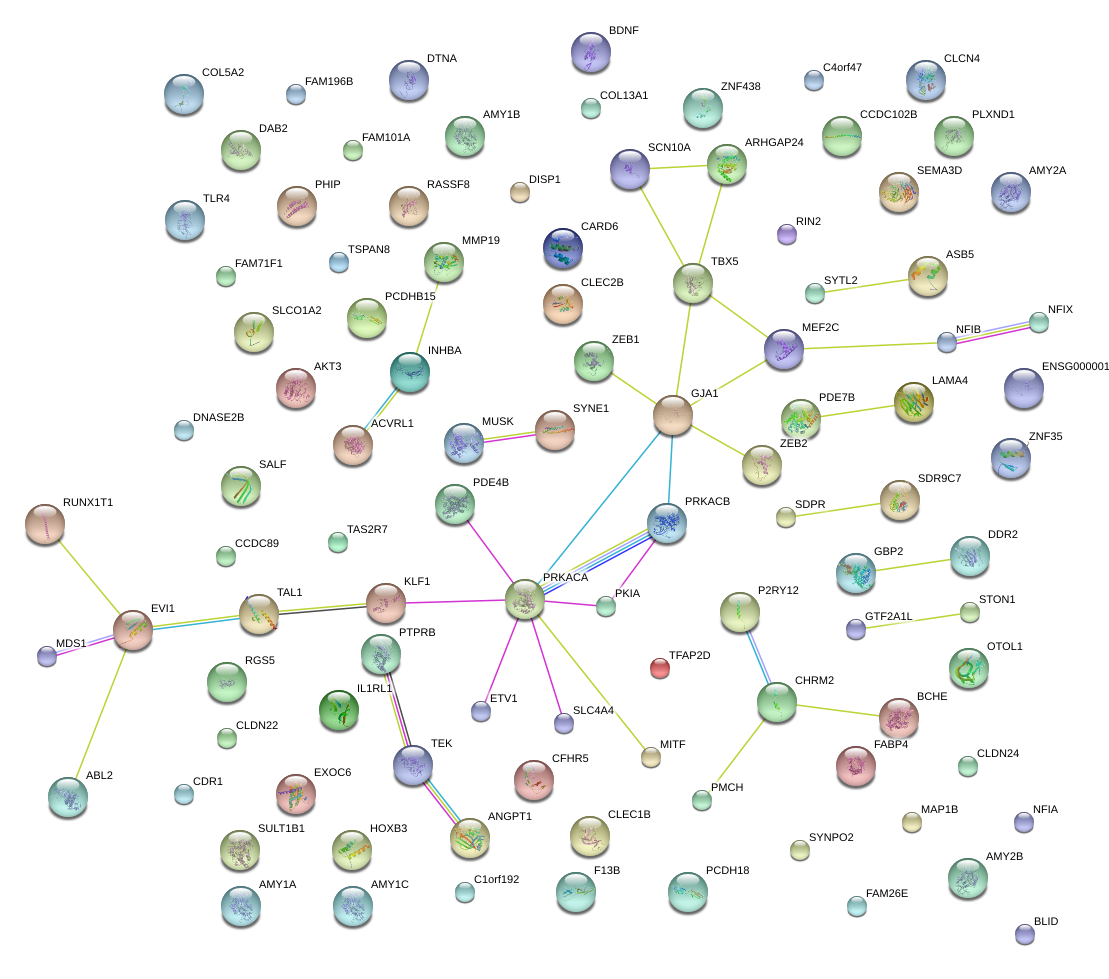
|
chr5_+_71538456
|
1.384
|
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr6_+_116939500
|
1.362
|
NM_153711
|
FAM26E
|
family with sequence similarity 26, member E
|
|
chr4_-_177427252
|
1.224
|
NM_080874
|
ASB5
|
ankyrin repeat and SOCS box containing 5
|
|
chr5_+_40877053
|
1.210
|
NM_032587
|
CARD6
|
caspase recruitment domain family, member 6
|
|
chr12_-_69317441
|
1.095
|
NM_001109754
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B
|
|
chr8_-_93176618
|
1.037
|
NM_001198629
NM_001198630
NM_001198632
NM_175635
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr1_-_161439487
|
1.025
|
NM_003617
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr11_-_121492010
|
0.972
|
NM_001001786
|
BLID
|
BH3-like motif containing, cell death inducer
|
|
chr2_-_144991386
|
0.936
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr4_+_86968069
|
0.925
|
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr20_+_19863716
|
0.906
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chr10_-_31328426
|
0.888
|
NM_001143769
|
ZNF438
|
zinc finger protein 438
|
|
chr7_+_128136347
|
0.875
|
|
FAM71F1
|
family with sequence similarity 71, member F1
|
|
chr1_+_66570363
|
0.815
|
NM_001037339
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific
|
|
chr9_+_112470871
|
0.814
|
NM_001166280
NM_001166281
NM_005592
|
MUSK
|
muscle, skeletal, receptor tyrosine kinase
|
|
chr1_+_84402962
|
0.783
|
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr4_-_70661018
|
0.773
|
NM_014465
|
SULT1B1
|
sulfotransferase family, cytosolic, 1B, member 1
|
|
chr1_+_84646621
|
0.771
|
NM_058248
|
DNASE2B
|
deoxyribonuclease II beta
|
|
chr7_-_13992351
|
0.768
|
NM_001163150
NM_001163151
NM_001163152
|
ETV1
|
ets variant 1
|
|
chr7_-_41709191
|
0.764
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chr8_-_108579270
|
0.763
|
NM_001146
|
ANGPT1
|
angiopoietin 1
|
|
chr1_+_160868822
|
0.746
|
NM_001014796
NM_006182
|
DDR2
|
discoidin domain receptor tyrosine kinase 2
|
|
chr3_-_130761967
|
0.722
|
|
PLXND1
|
plexin D1
|
|
chr12_+_123339662
|
0.705
|
NM_181709
|
FAM101A
|
family with sequence similarity 101, member A
|
|
chr1_+_160868932
|
0.684
|
|
DDR2
|
discoidin domain receptor tyrosine kinase 2
|
|
chr12_-_21379098
|
0.678
|
NM_021094
|
SLCO1A2
|
solute carrier organic anion transporter family, member 1A2
|
|
chr6_-_112682459
|
0.676
|
NM_001105206
NM_001105207
NM_001105208
NM_001105209
NM_002290
|
LAMA4
|
laminin, alpha 4
|
|
chr1_-_89364273
|
0.671
|
NM_004120
|
GBP2
|
guanylate binding protein 2, interferon-inducible
|
|
chr12_+_50592379
|
0.668
|
NM_001077401
|
ACVRL1
|
activin A receptor type II-like 1
|
|
chr10_+_94584449
|
0.665
|
NM_001013848
|
EXOC6
|
exocyst complex component 6
|
|
chr11_-_27679617
|
0.650
|
NM_170733
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr8_-_93144366
|
0.644
|
NM_004349
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr2_+_102320148
|
0.643
|
NM_003856
|
IL1RL1
|
interleukin 1 receptor-like 1
|
|
chr6_-_112682401
|
0.637
|
|
LAMA4
|
laminin, alpha 4
|
|
chr6_-_152744022
|
0.636
|
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1
|
|
chr9_+_119506428
|
0.630
|
|
TLR4
|
toll-like receptor 4
|
|
chr4_-_138673078
|
0.630
|
NM_019035
|
PCDH18
|
protocadherin 18
|
|
chr2_-_189752575
|
0.621
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr4_+_186587537
|
0.620
|
NM_001114357
|
C4orf47
|
chromosome 4 open reading frame 47
|
|
chr5_-_169340321
|
0.593
|
NM_001129891
|
FAM196B
|
family with sequence similarity 196, member B
|
|
chr11_-_104274606
|
0.593
|
NM_001191016
|
CASP12
|
caspase 12 (gene/pseudogene)
|
|
chr8_-_93099040
|
0.593
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr2_-_144991556
|
0.575
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr12_+_26017954
|
0.573
|
NM_001164747
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8
|
|
chr11_-_85146691
|
0.563
|
NM_001162951
NM_001162953
|
SYTL2
|
synaptotagmin-like 2
|
|
chr1_-_177378801
|
0.559
|
NM_001136000
NM_001168239
NM_005158
|
ABL2
|
v-abl Abelson murine leukemia viral oncogene homolog 2
|
|
chr12_-_101115677
|
0.558
|
NM_002674
|
PMCH
|
pro-melanin-concentrating hormone
|
|
chr10_+_71231649
|
0.555
|
NM_001130103
NM_080798
NM_080800
NM_080801
NM_080802
NM_080805
|
COL13A1
|
collagen, type XIII, alpha 1
|
|
chr2_-_192419896
|
0.553
|
|
SDPR
|
serum deprivation response
|
|
chr12_-_69838045
|
0.552
|
NM_004616
|
TSPAN8
|
tetraspanin 8
|
|
chr6_+_136214495
|
0.549
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr9_-_3213007
|
0.542
|
|
|
|
|
chr8_-_93176819
|
0.540
|
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr9_+_27099138
|
0.538
|
NM_000459
|
TEK
|
TEK tyrosine kinase, endothelial
|
|
chr6_-_112682573
|
0.538
|
|
LAMA4
|
laminin, alpha 4
|
|
chr17_-_44022595
|
0.533
|
|
HOXB3
|
homeobox B3
|
|
chr12_-_69837831
|
0.531
|
|
TSPAN8
|
tetraspanin 8
|
|
chr5_-_88155360
|
0.521
|
NM_001193348
NM_001193349
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr9_+_119506274
|
0.519
|
NM_138554
|
TLR4
|
toll-like receptor 4
|
|
chr1_+_221168388
|
0.517
|
NM_032890
|
DISP1
|
dispatched homolog 1 (Drosophila)
|
|
chr8_-_93181091
|
0.510
|
NM_001198628
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr3_-_170346760
|
0.507
|
NM_001105077
NM_001163999
NM_005241
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr6_+_121798443
|
0.505
|
NM_000165
|
GJA1
|
gap junction protein, alpha 1, 43kDa
|
|
chr7_+_136203938
|
0.502
|
NM_001006627
NM_001006630
|
CHRM2
|
cholinergic receptor, muscarinic 2
|
|
chr3_-_152585226
|
0.498
|
NM_022788
|
P2RY12
|
purinergic receptor P2Y, G-protein coupled, 12
|
|
chr1_-_159604287
|
0.498
|
NM_001013625
|
C1orf192
|
chromosome 1 open reading frame 192
|
|
chrX_+_10084984
|
0.494
|
NM_001830
|
CLCN4
|
chloride channel 4
|
|
chr12_-_113330574
|
0.489
|
NM_000192
NM_080717
|
TBX5
|
T-box 5
|
|
chr6_+_79844267
|
0.488
|
|
PHIP
|
pleckstrin homology domain interacting protein
|
|
chr4_+_72423633
|
0.488
|
NM_003759
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr17_-_44006808
|
0.478
|
NM_002146
|
HOXB3
|
homeobox B3
|
|
chr4_-_184478920
|
0.473
|
NM_001111319
|
CLDN22
|
claudin 22
|
|
chr12_-_54517241
|
0.470
|
|
MMP19
|
matrix metallopeptidase 19
|
|
chr19_-_12858956
|
0.470
|
NM_006563
|
KLF1
|
Kruppel-like factor 1 (erythroid)
|
|
chr7_-_84589182
|
0.467
|
NM_152754
|
SEMA3D
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D
|
|
chr5_-_39460687
|
0.464
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr5_-_39430180
|
0.461
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chrX_-_139694248
|
0.460
|
NM_004065
|
CDR1
|
cerebellar degeneration-related protein 1, 34kDa
|
|
chr1_-_47468029
|
0.455
|
NM_003189
|
TAL1
|
T-cell acute lymphocytic leukemia 1
|
|
chr3_-_167037861
|
0.453
|
|
BCHE
|
butyrylcholinesterase
|
|
chr3_+_162697289
|
0.453
|
NM_001080440
|
OTOL1
|
otolin 1 homolog (zebrafish)
|
|
chr1_+_195213289
|
0.452
|
NM_030787
|
CFHR5
|
complement factor H-related 5
|
|
chr12_-_9913199
|
0.445
|
|
CLEC2B
|
C-type lectin domain family 2, member B
|
|
chr8_+_79591074
|
0.443
|
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha
|
|
chr3_+_70068440
|
0.441
|
NM_000248
NM_001184968
NM_198158
NM_198178
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr3_-_170347019
|
0.434
|
NM_001105078
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr8_-_82557956
|
0.433
|
NM_001442
|
FABP4
|
fatty acid binding protein 4, adipocyte
|
|
chr1_+_103999824
|
0.431
|
NM_004038
NM_001008218
NM_001008219
|
AMY1A
AMY1B
AMY1C
|
amylase, alpha 1A (salivary)
amylase, alpha 1B (salivary)
amylase, alpha 1C (salivary)
|
|
chr18_+_30589937
|
0.430
|
NM_001390
NM_001391
|
DTNA
|
dystrobrevin, alpha
|
|
chr18_+_64616296
|
0.427
|
NM_024781
|
CCDC102B
|
coiled-coil domain containing 102B
|
|
chr1_-_242073094
|
0.426
|
NM_005465
NM_181690
|
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 (protein kinase B, gamma)
|
|
chr11_-_27679022
|
0.426
|
NM_001143812
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr4_+_86615275
|
0.424
|
NM_001025616
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr2_+_48649662
|
0.421
|
NM_172311
|
STON1-GTF2A1L
|
STON1-GTF2A1L readthrough
|
|
chr3_+_44665220
|
0.420
|
NM_003420
|
ZNF35
|
zinc finger protein 35
|
|
chr1_-_195302973
|
0.414
|
NM_001994
|
F13B
|
coagulation factor XIII, B polypeptide
|
|
chr6_+_50789215
|
0.412
|
NM_172238
|
TFAP2D
|
transcription factor AP-2 delta (activating enhancer binding protein 2 delta)
|
|
chr12_-_55614311
|
0.398
|
NM_148897
|
SDR9C7
|
short chain dehydrogenase/reductase family 9C, member 7
|
|
chr4_+_120167264
|
0.392
|
|
SYNPO2
|
synaptopodin 2
|
|
chr9_-_14076901
|
0.389
|
|
NFIB
|
nuclear factor I/B
|
|
chr3_-_38810504
|
0.388
|
NM_006514
|
SCN10A
|
sodium channel, voltage-gated, type X, alpha subunit
|
|
chr11_-_85074848
|
0.387
|
NM_152723
|
CCDC89
|
coiled-coil domain containing 89
|
|
chr12_-_10846435
|
0.382
|
NM_023919
|
TAS2R7
|
taste receptor, type 2, member 7
|
|
chr5_+_140605100
|
0.382
|
|
PCDHB15
|
protocadherin beta 15
|
|
chr1_+_170656450
|
0.382
|
NM_139240
|
C1orf105
|
chromosome 1 open reading frame 105
|
|
chr6_-_128281434
|
0.380
|
NM_001164687
|
THEMIS
|
thymocyte selection associated
|
|
chr4_+_120276386
|
0.379
|
NM_016599
|
MYOZ2
|
myozenin 2
|
|
chr4_+_111616630
|
0.378
|
NM_001977
|
ENPEP
|
glutamyl aminopeptidase (aminopeptidase A)
|
|
chr14_+_22915772
|
0.376
|
NM_001037288
NM_138460
|
CMTM5
|
CKLF-like MARVEL transmembrane domain containing 5
|
|
chr5_-_13997588
|
0.376
|
NM_001369
|
DNAH5
|
dynein, axonemal, heavy chain 5
|
|
chr4_-_70861301
|
0.375
|
NM_001891
|
CSN2
|
casein beta
|
|
chr2_+_210152399
|
0.373
|
|
MAP2
|
microtubule-associated protein 2
|
|
chr1_+_92405196
|
0.372
|
NM_015237
|
KIAA1107
|
KIAA1107
|
|
chr7_-_13997574
|
0.371
|
NM_001163148
|
ETV1
|
ets variant 1
|
|
chr12_+_79362256
|
0.370
|
NM_001145026
|
PTPRQ
|
protein tyrosine phosphatase, receptor type, Q
|
|
chrX_-_10605726
|
0.366
|
NM_001098624
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr11_+_10433241
|
0.363
|
NM_001025389
|
AMPD3
|
adenosine monophosphate deaminase 3
|
|
chr11_+_72697310
|
0.359
|
NM_014786
|
ARHGEF17
|
Rho guanine nucleotide exchange factor (GEF) 17
|
|
chr3_-_168853982
|
0.359
|
NM_178824
|
WDR49
|
WD repeat domain 49
|
|
chr5_+_140552087
|
0.358
|
NM_018930
|
PCDHB10
|
protocadherin beta 10
|
|
chr4_+_72321113
|
0.358
|
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr4_-_152316105
|
0.354
|
NM_001128924
|
SH3D19
|
SH3 domain containing 19
|
|
chr3_+_44667504
|
0.354
|
|
ZNF35
|
zinc finger protein 35
|
|
chr18_+_41560915
|
0.350
|
NM_001146037
|
SLC14A1
|
solute carrier family 14 (urea transporter), member 1 (Kidd blood group)
|
|
chr2_-_160838397
|
0.350
|
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr15_-_70350609
|
0.349
|
NM_020214
|
PARP6
|
poly (ADP-ribose) polymerase family, member 6
|
|
chr1_+_148389039
|
0.347
|
|
PLEKHO1
|
pleckstrin homology domain containing, family O member 1
|
|
chr1_+_160002733
|
0.346
|
|
ATF6
|
activating transcription factor 6
|
|
chr16_+_75790849
|
0.344
|
NM_001129979
|
SYCE1L
|
synaptonemal complex central element protein 1-like
|
|
chr7_-_13997389
|
0.343
|
NM_001163149
|
ETV1
|
ets variant 1
|
|
chr16_+_82540172
|
0.343
|
NM_013370
NM_182980
|
OSGIN1
|
oxidative stress induced growth inhibitor 1
|
|
chr2_-_192420168
|
0.335
|
NM_004657
|
SDPR
|
serum deprivation response
|
|
chr17_-_59817456
|
0.334
|
|
PECAM1
|
platelet/endothelial cell adhesion molecule
|
|
chrX_+_71280758
|
0.334
|
NM_207422
|
FLJ44635
|
TPT1-like protein
|
|
chr4_+_88939725
|
0.333
|
NM_004967
|
IBSP
|
integrin-binding sialoprotein
|
|
chr14_+_95792299
|
0.332
|
NM_000710
|
BDKRB1
|
bradykinin receptor B1
|
|
chr9_-_35802142
|
0.332
|
NM_001039592
NM_172312
|
SPAG8
|
sperm associated antigen 8
|
|
chr19_+_49147219
|
0.332
|
NM_013359
|
ZNF221
|
zinc finger protein 221
|
|
chr12_+_6751930
|
0.330
|
NM_002286
|
LAG3
|
lymphocyte-activation gene 3
|
|
chr5_+_140181405
|
0.327
|
NM_018908
NM_031501
|
PCDHA5
|
protocadherin alpha 5
|
|
chr1_-_34404029
|
0.327
|
NM_052896
|
CSMD2
|
CUB and Sushi multiple domains 2
|
|
chr1_+_203106600
|
0.327
|
|
NFASC
|
neurofascin
|
|
chr3_+_11242666
|
0.326
|
NM_001098211
|
HRH1
|
histamine receptor H1
|
|
chr5_+_92944680
|
0.326
|
NM_005654
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr7_-_27186400
|
0.322
|
NM_153715
|
HOXA10
|
homeobox A10
|
|
chr1_-_161439248
|
0.321
|
NM_001195303
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr8_+_79590890
|
0.320
|
NM_006823
NM_181839
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha
|
|
chr15_+_73803351
|
0.320
|
NM_175881
|
ODF3L1
|
outer dense fiber of sperm tails 3-like 1
|
|
chr11_-_85103438
|
0.320
|
NM_032379
|
SYTL2
|
synaptotagmin-like 2
|
|
chr8_-_15140162
|
0.319
|
NM_139167
|
SGCZ
|
sarcoglycan, zeta
|
|
chr11_+_36546138
|
0.319
|
NM_000448
|
RAG1
|
recombination activating gene 1
|
|
chr11_-_51269023
|
0.318
|
NM_001005272
|
OR4A5
|
olfactory receptor, family 4, subfamily A, member 5
|
|
chr3_-_55489973
|
0.315
|
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr21_+_44818033
|
0.313
|
NM_198687
|
KRTAP10-4
|
keratin associated protein 10-4
|
|
chr19_+_53450743
|
0.313
|
|
|
|
|
chr2_-_133895795
|
0.312
|
|
NCKAP5
|
NCK-associated protein 5
|
|
chr3_+_153014550
|
0.312
|
NM_001086
|
AADAC
|
arylacetamide deacetylase (esterase)
|
|
chr12_+_79127363
|
0.311
|
NM_173591
|
OTOGL
|
otogelin-like
|
|
chr22_-_37211467
|
0.311
|
|
DDX17
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 17
|
|
chr19_+_13767207
|
0.310
|
NM_023072
|
ZSWIM4
|
zinc finger, SWIM-type containing 4
|
|
chr3_+_123996560
|
0.310
|
NM_032839
|
DIRC2
|
disrupted in renal carcinoma 2
|
|
chr17_+_26322168
|
0.310
|
|
RNF135
|
ring finger protein 135
|
|
chr11_-_55518675
|
0.309
|
NM_003697
|
OR5F1
|
olfactory receptor, family 5, subfamily F, member 1
|
|
chr2_-_151854619
|
0.308
|
NM_004688
|
NMI
|
N-myc (and STAT) interactor
|
|
chr21_+_29439768
|
0.308
|
|
C21orf7
|
chromosome 21 open reading frame 7
|
|
chr12_+_46799279
|
0.306
|
NM_000289
|
PFKM
|
phosphofructokinase, muscle
|
|
chrX_-_50403412
|
0.305
|
|
SHROOM4
|
shroom family member 4
|
|
chr5_+_55098469
|
0.304
|
NM_001166534
|
DDX4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4
|
|
chr3_+_108578877
|
0.303
|
NM_032600
|
CCDC54
|
coiled-coil domain containing 54
|
|
chr13_+_22653059
|
0.302
|
NM_000231
|
SGCG
|
sarcoglycan, gamma (35kDa dystrophin-associated glycoprotein)
|
|
chr4_+_56731117
|
0.302
|
NM_020722
|
KIAA1211
|
KIAA1211
|
|
chr1_+_68070573
|
0.302
|
|
|
|
|
chr13_-_109600709
|
0.301
|
|
COL4A1
|
collagen, type IV, alpha 1
|
|
chr7_+_28305464
|
0.300
|
NM_182899
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr3_-_15357878
|
0.299
|
NM_001018009
|
SH3BP5
|
SH3-domain binding protein 5 (BTK-associated)
|
|
chr4_-_77176163
|
0.297
|
NM_005409
|
CXCL11
|
chemokine (C-X-C motif) ligand 11
|
|
chr15_+_19869841
|
0.297
|
NM_001004719
|
OR4M2
|
olfactory receptor, family 4, subfamily M, member 2
|
|
chr9_+_74956494
|
0.297
|
NM_000700
|
ANXA1
|
annexin A1
|
|
chr9_+_117955890
|
0.295
|
NM_002581
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1
|
|
chr1_-_31117948
|
0.294
|
|
SDC3
|
syndecan 3
|
|
chr20_-_62058162
|
0.294
|
NM_017859
|
UCKL1
|
uridine-cytidine kinase 1-like 1
|
|
chr10_+_71231693
|
0.293
|
|
COL13A1
|
collagen, type XIII, alpha 1
|
|
chr16_+_54158084
|
0.292
|
NM_032330
|
CAPNS2
|
calpain, small subunit 2
|
|
chr11_+_55174953
|
0.292
|
NM_001004059
|
OR4S2
|
olfactory receptor, family 4, subfamily S, member 2
|
|
chr2_+_210152609
|
0.292
|
NM_002374
NM_031845
NM_031847
|
MAP2
|
microtubule-associated protein 2
|
|
chr10_-_115413630
|
0.292
|
NM_006175
NM_198060
|
NRAP
|
nebulin-related anchoring protein
|
|
chr10_+_124141808
|
0.291
|
NM_021622
|
PLEKHA1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1
|
|
chr8_-_13018123
|
0.290
|
NM_001164271
|
DLC1
|
deleted in liver cancer 1
|
|
chr15_+_47502666
|
0.290
|
NM_002009
|
FGF7
|
fibroblast growth factor 7
|
|
chr19_+_18357966
|
0.288
|
NM_004864
|
GDF15
|
growth differentiation factor 15
|
|
chr10_-_91285290
|
0.285
|
NM_213606
|
SLC16A12
|
solute carrier family 16, member 12 (monocarboxylic acid transporter 12)
|
|
chr8_-_93177057
|
0.282
|
NM_001198625
NM_001198626
NM_001198627
NM_001198631
NM_001198634
NM_001198679
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr12_+_32578513
|
0.282
|
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr3_-_158700036
|
0.280
|
NM_024621
|
VEPH1
|
ventricular zone expressed PH domain homolog 1 (zebrafish)
|
|
chr7_-_87694218
|
0.280
|
NM_198901
|
SRI
|
sorcin
|
|
chr7_-_15568099
|
0.278
|
NM_001004320
|
AGMO
|
alkylglycerol monooxygenase
|
|
chr6_-_66346417
|
0.275
|
NM_198283
|
EYS
|
eyes shut homolog (Drosophila)
|
|
chr2_+_66519949
|
0.273
|
|
MEIS1
|
Meis homeobox 1
|
|
chr12_-_21980894
|
0.273
|
NM_005691
NM_020297
NM_020298
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9
|
|
chr9_-_35553895
|
0.272
|
NM_001099951
NM_001164310
|
FAM166B
|
family with sequence similarity 166, member B
|