|
chr19_+_61742111
|
2.042
|
NM_020828
|
ZFP28
|
zinc finger protein 28 homolog (mouse)
|
|
chr19_-_50691999
|
1.936
|
NM_005619
NM_206900
|
RTN2
|
reticulon 2
|
|
chr17_+_45488330
|
1.824
|
NM_002204
NM_005501
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor)
|
|
chr11_-_94603857
|
1.730
|
NM_144665
|
SESN3
|
sestrin 3
|
|
chr11_+_65990869
|
1.640
|
NM_001098510
NM_145065
|
PELI3
|
pellino homolog 3 (Drosophila)
|
|
chr19_+_5855842
|
1.600
|
NM_001017921
|
VMAC
|
vimentin-type intermediate filament associated coiled-coil protein
|
|
chrX_-_149817785
|
1.565
|
NM_001184808
NM_031462
NM_134445
NM_134446
|
CD99L2
|
CD99 molecule-like 2
|
|
chr2_-_37752272
|
1.515
|
|
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3
|
|
chr17_+_45488356
|
1.490
|
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor)
|
|
chr19_-_54835162
|
1.386
|
NM_006270
|
RRAS
|
related RAS viral (r-ras) oncogene homolog
|
|
chr3_+_137167296
|
1.385
|
|
PPP2R3A
|
protein phosphatase 2, regulatory subunit B'', alpha
|
|
chrX_-_135161185
|
1.371
|
NM_001173517
NM_024597
|
MAP7D3
|
MAP7 domain containing 3
|
|
chr19_+_45976001
|
1.358
|
NM_016154
|
RAB4B-EGLN2
RAB4B
|
RAB4B-EGLN2 read-through transcript
RAB4B, member RAS oncogene family
|
|
chr11_-_65443015
|
1.347
|
NM_001135635
NM_031450
|
C11orf68
|
chromosome 11 open reading frame 68
|
|
chr19_-_45888259
|
1.312
|
NM_004756
|
NUMBL
|
numb homolog (Drosophila)-like
|
|
chr2_-_235070431
|
1.250
|
NM_005737
|
ARL4C
|
ADP-ribosylation factor-like 4C
|
|
chr11_+_65443303
|
1.225
|
NM_006442
|
DRAP1
|
DR1-associated protein 1 (negative cofactor 2 alpha)
|
|
chr5_-_2804768
|
1.203
|
NM_001134222
NM_033267
|
IRX2
|
iroquois homeobox 2
|
|
chr14_+_95928189
|
1.193
|
NM_152327
|
AK7
|
adenylate kinase 7
|
|
chr8_-_144887876
|
1.183
|
NM_198488
|
FAM83H
|
family with sequence similarity 83, member H
|
|
chr15_+_61356801
|
1.170
|
NM_001145646
NM_031301
|
APH1B
|
anterior pharynx defective 1 homolog B (C. elegans)
|
|
chr2_-_37752731
|
1.152
|
NM_006449
|
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3
|
|
chr15_-_70399301
|
1.150
|
NM_001172684
|
CELF6
|
CUGBP, Elav-like family member 6
|
|
chr20_-_56317894
|
1.137
|
|
PPP4R1L
|
protein phosphatase 4, regulatory subunit 1-like
|
|
chr10_+_11824361
|
1.110
|
NM_024693
|
ECHDC3
|
enoyl CoA hydratase domain containing 3
|
|
chr12_+_48303607
|
1.106
|
NM_012272
|
PRPF40B
|
PRP40 pre-mRNA processing factor 40 homolog B (S. cerevisiae)
|
|
chr19_-_12747263
|
1.099
|
|
HOOK2
|
hook homolog 2 (Drosophila)
|
|
chr13_+_41520888
|
1.082
|
NM_152910
NM_178009
|
DGKH
|
diacylglycerol kinase, eta
|
|
chr13_-_109236897
|
1.056
|
NM_003749
|
IRS2
|
insulin receptor substrate 2
|
|
chrX_+_64804235
|
1.054
|
NM_002444
|
MSN
|
moesin
|
|
chr10_-_134940315
|
1.052
|
NM_001109
NM_001164489
NM_001164490
|
ADAM8
|
ADAM metallopeptidase domain 8
|
|
chr19_-_15204767
|
1.048
|
NM_001142886
|
EPHX3
|
epoxide hydrolase 3
|
|
chr6_-_31805547
|
1.047
|
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2
|
|
chr19_+_10574055
|
1.046
|
NM_001145056
|
SLC44A2
|
solute carrier family 44, member 2
|
|
chrX_+_64804259
|
1.041
|
|
MSN
|
moesin
|
|
chr5_-_16989290
|
1.039
|
|
MYO10
|
myosin X
|
|
chr22_-_29015452
|
1.019
|
NM_001037666
|
GATSL3
|
GATS protein-like 3
|
|
chr1_-_113059396
|
1.011
|
NM_005167
|
PPM1J
|
protein phosphatase, Mg2+/Mn2+ dependent, 1J
|
|
chrX_+_64804318
|
1.005
|
|
MSN
|
moesin
|
|
chr6_-_29708647
|
1.004
|
|
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1
|
|
chr1_+_32992131
|
1.004
|
|
|
|
|
chr8_-_22044463
|
0.991
|
NM_005144
NM_018411
|
HR
|
hairless homolog (mouse)
|
|
chr19_-_6718417
|
0.967
|
NM_005490
|
SH2D3A
|
SH2 domain containing 3A
|
|
chr22_-_29052863
|
0.953
|
NM_031937
|
TBC1D10A
|
TBC1 domain family, member 10A
|
|
chr12_+_56406296
|
0.943
|
|
LOC100130776
|
hypothetical LOC100130776
|
|
chr19_+_60487345
|
0.933
|
NM_032430
|
BRSK1
|
BR serine/threonine kinase 1
|
|
chr12_-_53099203
|
0.933
|
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr12_-_45759691
|
0.932
|
NM_001143668
NM_181847
|
AMIGO2
|
adhesion molecule with Ig-like domain 2
|
|
chr10_-_105604942
|
0.931
|
NM_014631
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr22_-_49050847
|
0.927
|
NM_002751
|
MAPK11
|
mitogen-activated protein kinase 11
|
|
chr12_-_53099277
|
0.923
|
NM_002205
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr1_-_6243621
|
0.923
|
NM_207370
|
GPR153
|
G protein-coupled receptor 153
|
|
chr19_-_19599973
|
0.912
|
NM_004720
|
LPAR2
|
lysophosphatidic acid receptor 2
|
|
chr3_+_185762233
|
0.911
|
NM_004443
|
EPHB3
|
EPH receptor B3
|
|
chr11_-_62146144
|
0.910
|
|
B3GAT3
|
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I)
|
|
chr19_+_18608827
|
0.909
|
NM_018316
|
KLHL26
|
kelch-like 26 (Drosophila)
|
|
chr11_-_11987115
|
0.908
|
|
DKK3
|
dickkopf homolog 3 (Xenopus laevis)
|
|
chr14_-_104515738
|
0.900
|
NM_138420
|
AHNAK2
|
AHNAK nucleoprotein 2
|
|
chrX_-_149817720
|
0.898
|
|
CD99L2
|
CD99 molecule-like 2
|
|
chr19_-_48976852
|
0.897
|
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4
|
|
chr8_-_144722141
|
0.886
|
|
C8orf73
|
chromosome 8 open reading frame 73
|
|
chrX_+_152891184
|
0.885
|
NM_003492
|
TMEM187
|
transmembrane protein 187
|
|
chr9_-_100057313
|
0.884
|
|
TBC1D2
|
TBC1 domain family, member 2
|
|
chr12_-_53099269
|
0.883
|
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr12_-_51900388
|
0.876
|
NM_001042728
|
RARG
|
retinoic acid receptor, gamma
|
|
chr11_-_127897271
|
0.871
|
NM_001162422
NM_005238
|
ETS1
|
v-ets erythroblastosis virus E26 oncogene homolog 1 (avian)
|
|
chr5_+_176006993
|
0.867
|
NM_001006616
NM_012171
NM_130465
|
TSPAN17
|
tetraspanin 17
|
|
chr1_+_156229686
|
0.864
|
NM_018240
|
KIRREL
|
kin of IRRE like (Drosophila)
|
|
chr9_+_129962292
|
0.863
|
NM_024112
|
C9orf16
|
chromosome 9 open reading frame 16
|
|
chr7_+_100584397
|
0.857
|
NM_001283
|
AP1S1
|
adaptor-related protein complex 1, sigma 1 subunit
|
|
chr1_-_46855116
|
0.857
|
NM_201403
|
MOBKL2C
|
MOB1, Mps One Binder kinase activator-like 2C (yeast)
|
|
chr8_+_32525269
|
0.856
|
NM_001160002
NM_001160004
NM_001160005
NM_001160007
NM_001160008
NM_004495
NM_013956
NM_013957
NM_013958
NM_013960
NM_013964
|
NRG1
|
neuregulin 1
|
|
chr9_+_139006663
|
0.854
|
NM_183241
|
C9orf142
|
chromosome 9 open reading frame 142
|
|
chrX_-_134060329
|
0.846
|
|
NCRNA00087
|
non-protein coding RNA 87
|
|
chr5_+_176007095
|
0.840
|
|
TSPAN17
|
tetraspanin 17
|
|
chr2_+_84986273
|
0.838
|
NM_021103
|
TMSB10
|
thymosin beta 10
|
|
chr5_+_176492662
|
0.837
|
NM_172349
|
NSD1
|
nuclear receptor binding SET domain protein 1
|
|
chr9_-_109291575
|
0.835
|
|
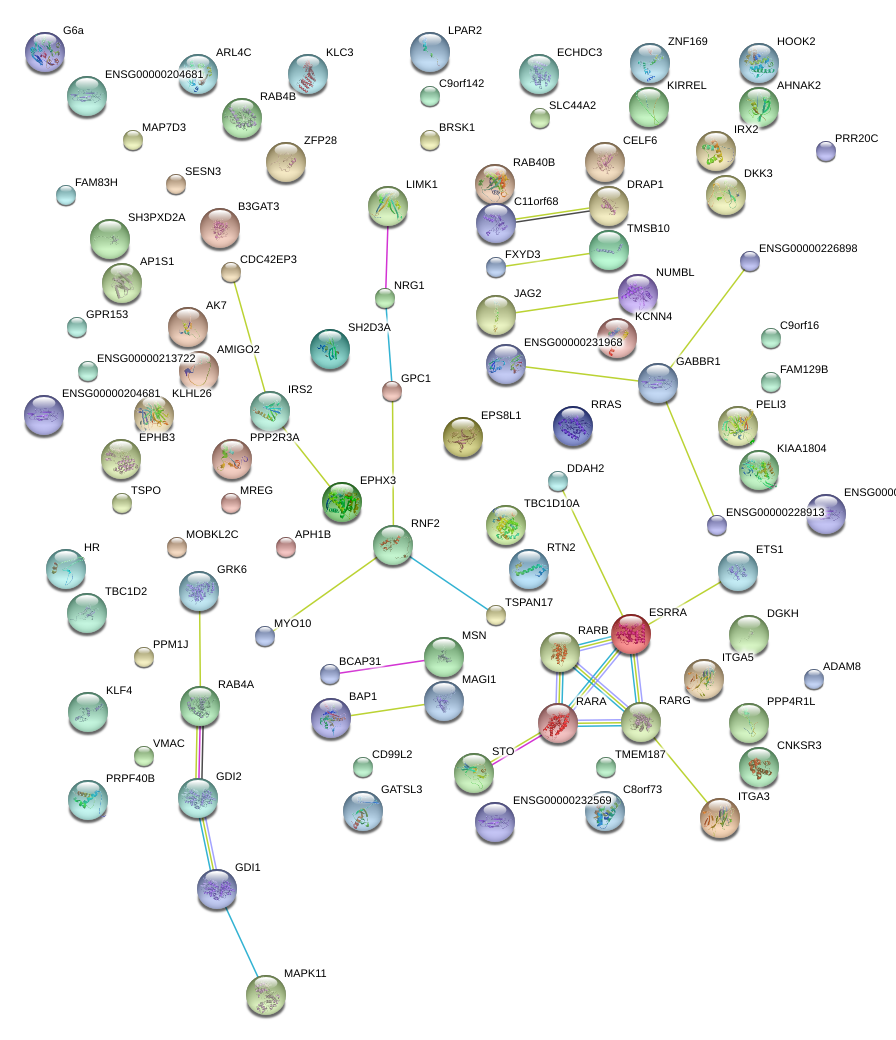
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr1_+_231530136
|
0.823
|
NM_032435
|
KIAA1804
|
mixed lineage kinase 4
|
|
chrX_-_152642716
|
0.820
|
NM_001139457
NM_001139441
|
BCAP31
|
B-cell receptor-associated protein 31
|
|
chr22_+_41877502
|
0.815
|
|
TSPO
|
translocator protein (18kDa)
|
|
chr11_+_63829616
|
0.814
|
NM_004451
|
ESRRA
|
estrogen-related receptor alpha
|
|
chr11_+_65443452
|
0.804
|
|
DRAP1
|
DR1-associated protein 1 (negative cofactor 2 alpha)
|
|
chr19_-_48977247
|
0.804
|
NM_002250
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4
|
|
chrX_+_153318452
|
0.799
|
NM_001493
|
GDI1
|
GDP dissociation inhibitor 1
|
|
chr9_-_129371171
|
0.793
|
NM_022833
|
FAM129B
|
family with sequence similarity 129, member B
|
|
chr5_+_176786279
|
0.790
|
NM_001004105
NM_001004106
NM_002082
|
GRK6
|
G protein-coupled receptor kinase 6
|
|
chr19_+_50535837
|
0.789
|
NM_177417
|
KLC3
|
kinesin light chain 3
|
|
chr19_+_60283571
|
0.788
|
NM_017729
|
EPS8L1
|
EPS8-like 1
|
|
chr2_+_241023908
|
0.786
|
|
GPC1
|
glypican 1
|
|
chr14_-_104705753
|
0.781
|
|
JAG2
|
jagged 2
|
|
chr19_+_40298571
|
0.776
|
NM_001136007
NM_001136008
NM_001136009
NM_001136010
NM_001136011
NM_001136012
NM_005971
NM_021910
|
FXYD3
|
FXYD domain containing ion transport regulator 3
|
|
chr17_-_78249750
|
0.775
|
NM_006822
|
RAB40B
|
RAB40B, member RAS oncogene family
|
|
chr3_-_52418933
|
0.774
|
NM_004656
|
BAP1
|
BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase)
|
|
chr7_+_73136085
|
0.771
|
NM_002314
|
LIMK1
|
LIM domain kinase 1
|
|
chr19_+_50196526
|
0.769
|
NM_006509
|
RELB
|
v-rel reticuloendotheliosis viral oncogene homolog B
|
|
chr11_+_65443484
|
0.766
|
|
DRAP1
|
DR1-associated protein 1 (negative cofactor 2 alpha)
|
|
chr19_-_2672337
|
0.766
|
NM_145173
|
DIRAS1
|
DIRAS family, GTP-binding RAS-like 1
|
|
chr19_-_18493886
|
0.765
|
NM_006532
|
ELL
|
elongation factor RNA polymerase II
|
|
chr19_+_45811316
|
0.762
|
|
LTBP4
|
latent transforming growth factor beta binding protein 4
|
|
chr11_-_525532
|
0.760
|
NM_001130442
NM_005343
NM_176795
|
HRAS
|
v-Ha-ras Harvey rat sarcoma viral oncogene homolog
|
|
chr19_-_18493847
|
0.751
|
|
ELL
|
elongation factor RNA polymerase II
|
|
chr18_-_50004770
|
0.747
|
|
MBD2
|
methyl-CpG binding domain protein 2
|
|
chrX_-_152643001
|
0.744
|
|
|
|
|
chr1_+_110554836
|
0.732
|
NM_001039574
NM_004978
|
KCNC4
|
potassium voltage-gated channel, Shaw-related subfamily, member 4
|
|
chr9_-_138560058
|
0.730
|
NM_017617
|
NOTCH1
|
notch 1
|
|
chr6_-_42527760
|
0.727
|
NM_033502
|
TRERF1
|
transcriptional regulating factor 1
|
|
chr14_+_104244998
|
0.727
|
|
INF2
|
inverted formin, FH2 and WH2 domain containing
|
|
chr19_-_45663463
|
0.723
|
NM_000713
|
BLVRB
|
biliverdin reductase B (flavin reductase (NADPH))
|
|
chrX_-_152517629
|
0.723
|
|
FAM58A
|
family with sequence similarity 58, member A
|
|
chr7_+_100302885
|
0.722
|
NM_003302
|
TRIP6
|
thyroid hormone receptor interactor 6
|
|
chr9_-_129370954
|
0.718
|
|
FAM129B
|
family with sequence similarity 129, member B
|
|
chr12_-_46585032
|
0.716
|
NM_000376
NM_001017535
|
VDR
|
vitamin D (1,25- dihydroxyvitamin D3) receptor
|
|
chrX_-_152643042
|
0.710
|
|
BCAP31
|
B-cell receptor-associated protein 31
|
|
chr3_+_128874458
|
0.696
|
NM_172027
|
ABTB1
|
ankyrin repeat and BTB (POZ) domain containing 1
|
|
chr3_-_46710137
|
0.694
|
NM_001190707
NM_147129
|
ALS2CL
|
ALS2 C-terminal like
|
|
chr1_+_182622987
|
0.692
|
|
C1orf21
|
chromosome 1 open reading frame 21
|
|
chr17_+_39990124
|
0.691
|
NM_001466
|
FZD2
|
frizzled homolog 2 (Drosophila)
|
|
chr1_-_32940850
|
0.688
|
|
SYNC
|
syncoilin, intermediate filament protein
|
|
chr10_-_3204955
|
0.685
|
|
PITRM1
|
pitrilysin metallopeptidase 1
|
|
chr1_-_29321599
|
0.684
|
NM_001171868
|
TMEM200B
|
transmembrane protein 200B
|
|
chr11_-_56848859
|
0.683
|
NM_033396
|
TNKS1BP1
|
tankyrase 1 binding protein 1, 182kDa
|
|
chr11_-_11987458
|
0.679
|
NM_013253
|
DKK3
|
dickkopf homolog 3 (Xenopus laevis)
|
|
chrX_-_152643016
|
0.676
|
NM_005745
|
BCAP31
|
B-cell receptor-associated protein 31
|
|
chr11_+_76249550
|
0.675
|
NM_018367
|
ACER3
|
alkaline ceramidase 3
|
|
chr7_+_115926855
|
0.674
|
|
CAV2
|
caveolin 2
|
|
chr11_-_11987414
|
0.673
|
|
DKK3
|
dickkopf homolog 3 (Xenopus laevis)
|
|
chr18_-_50004932
|
0.673
|
|
MBD2
|
methyl-CpG binding domain protein 2
|
|
chr8_+_144888275
|
0.669
|
|
LOC100128338
|
hypothetical LOC100128338
|
|
chr4_+_87734489
|
0.668
|
NM_006264
NM_080683
NM_080684
NM_080685
|
PTPN13
|
protein tyrosine phosphatase, non-receptor type 13 (APO-1/CD95 (Fas)-associated phosphatase)
|
|
chr9_+_139619918
|
0.668
|
|
ARRDC1
|
arrestin domain containing 1
|
|
chr18_+_32131628
|
0.667
|
NM_025135
|
FHOD3
|
formin homology 2 domain containing 3
|
|
chr1_+_149750485
|
0.667
|
NM_020770
|
CGN
|
cingulin
|
|
chrX_-_152517815
|
0.667
|
NM_001130997
NM_152274
|
FAM58A
|
family with sequence similarity 58, member A
|
|
chr1_-_27211913
|
0.665
|
NM_052943
|
FAM46B
|
family with sequence similarity 46, member B
|
|
chr5_+_10617431
|
0.664
|
NM_001164440
|
ANKRD33B
|
ankyrin repeat domain 33B
|
|
chr12_-_47868563
|
0.662
|
|
TUBA1A
|
tubulin, alpha 1a
|
|
chr17_-_7323528
|
0.658
|
|
ZBTB4
|
zinc finger and BTB domain containing 4
|
|
chr22_-_17489718
|
0.657
|
|
DGCR2
|
DiGeorge syndrome critical region gene 2
|
|
chr12_+_115455579
|
0.654
|
|
NCRNA00173
|
non-protein coding RNA 173
|
|
chr21_+_34367747
|
0.653
|
|
MRPS6
|
mitochondrial ribosomal protein S6
|
|
chr7_-_72791090
|
0.651
|
NM_001145364
NM_148912
NM_148913
|
ABHD11
|
abhydrolase domain containing 11
|
|
chr11_-_11987198
|
0.651
|
NM_015881
|
DKK3
|
dickkopf homolog 3 (Xenopus laevis)
|
|
chr9_+_35819375
|
0.651
|
NM_001042590
|
TMEM8B
|
transmembrane protein 8B
|
|
chr19_+_7801110
|
0.651
|
NM_145245
|
EVI5L
|
ecotropic viral integration site 5-like
|
|
chr3_-_52419056
|
0.651
|
|
BAP1
|
BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase)
|
|
chr15_+_39907447
|
0.649
|
NM_001114632
NM_001198588
NM_005090
|
JMJD7
JMJD7-PLA2G4B
|
jumonji domain containing 7
JMJD7-PLA2G4B readthrough
|
|
chr11_-_66877519
|
0.647
|
NM_021173
|
POLD4
|
polymerase (DNA-directed), delta 4
|
|
chr10_+_112247614
|
0.637
|
NM_004419
|
DUSP5
|
dual specificity phosphatase 5
|
|
chr2_+_241023703
|
0.633
|
NM_002081
|
GPC1
|
glypican 1
|
|
chr11_+_56984702
|
0.632
|
NM_178570
|
RTN4RL2
|
reticulon 4 receptor-like 2
|
|
chr9_-_35101534
|
0.630
|
|
KIAA1539
|
KIAA1539
|
|
chr4_+_79691875
|
0.627
|
|
ANXA3
|
annexin A3
|
|
chr22_+_36632242
|
0.626
|
NM_033386
|
MICALL1
|
MICAL-like 1
|
|
chr18_+_11971457
|
0.622
|
|
IMPA2
|
inositol(myo)-1(or 4)-monophosphatase 2
|
|
chr8_-_145088679
|
0.621
|
NM_201383
|
PLEC
|
plectin
|
|
chr9_-_130830255
|
0.620
|
NM_020145
|
SH3GLB2
|
SH3-domain GRB2-like endophilin B2
|
|
chr17_-_73867748
|
0.619
|
NM_003955
|
SOCS3
|
suppressor of cytokine signaling 3
|
|
chrX_+_117513899
|
0.617
|
NM_144658
|
DOCK11
|
dedicator of cytokinesis 11
|
|
chr3_-_52065130
|
0.617
|
NM_001947
|
DUSP7
|
dual specificity phosphatase 7
|
|
chr9_+_139619969
|
0.614
|
|
ARRDC1
|
arrestin domain containing 1
|
|
chr11_+_64838882
|
0.612
|
NM_006779
|
CDC42EP2
|
CDC42 effector protein (Rho GTPase binding) 2
|
|
chr22_-_37878394
|
0.611
|
NM_175709
|
CBX7
|
chromobox homolog 7
|
|
chr1_-_208046011
|
0.608
|
NM_006147
|
IRF6
|
interferon regulatory factor 6
|
|
chr8_+_144870482
|
0.606
|
NM_139021
|
MAPK15
|
mitogen-activated protein kinase 15
|
|
chr12_+_12935619
|
0.603
|
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A
|
|
chr9_+_138497767
|
0.602
|
NM_152571
|
C9orf163
|
chromosome 9 open reading frame 163
|
|
chrX_+_17303458
|
0.599
|
NM_198270
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies)
|
|
chr1_-_11788550
|
0.599
|
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H)
|
|
chr3_-_48607596
|
0.598
|
NM_000094
|
COL7A1
|
collagen, type VII, alpha 1
|
|
chr17_-_40674883
|
0.597
|
|
|
|
|
chr8_-_145122884
|
0.596
|
NM_000445
|
PLEC
|
plectin
|
|
chr21_+_46568460
|
0.595
|
NM_006031
|
PCNT
|
pericentrin
|
|
chr8_-_127639647
|
0.591
|
|
FAM84B
|
family with sequence similarity 84, member B
|
|
chr12_-_48508411
|
0.590
|
NM_001037806
|
NCKAP5L
|
NCK-associated protein 5-like
|
|
chr19_+_53819460
|
0.589
|
|
SPHK2
|
sphingosine kinase 2
|
|
chr1_-_33420213
|
0.585
|
NM_018207
|
TRIM62
|
tripartite motif containing 62
|
|
chrX_+_48915127
|
0.584
|
NM_002668
|
PLP2
|
proteolipid protein 2 (colonic epithelium-enriched)
|
|
chr7_-_87687302
|
0.581
|
NM_003130
|
SRI
|
sorcin
|
|
chr11_-_62145952
|
0.579
|
NM_012200
|
B3GAT3
|
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I)
|
|
chr19_+_54558798
|
0.578
|
NM_001197302
NM_001197301
NM_014419
|
DKKL1
|
dickkopf-like 1
|
|
chr2_-_1727292
|
0.577
|
NM_012293
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr10_-_116154504
|
0.576
|
NM_001001936
NM_032550
|
AFAP1L2
|
actin filament associated protein 1-like 2
|
|
chr12_+_56202874
|
0.576
|
NM_052897
|
MBD6
|
methyl-CpG binding domain protein 6
|
|
chr13_+_72531098
|
0.575
|
NM_001730
|
KLF5
|
Kruppel-like factor 5 (intestinal)
|
|
chr19_-_10807866
|
0.575
|
NM_006858
|
TMED1
|
transmembrane emp24 protein transport domain containing 1
|
|
chr8_+_22513043
|
0.574
|
NM_001013842
NM_001198827
NM_173686
|
C8orf58
|
chromosome 8 open reading frame 58
|
|
chr6_-_31805932
|
0.574
|
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2
|
|
chr9_+_35819208
|
0.573
|
NM_016446
NM_001042589
|
TMEM8B
|
transmembrane protein 8B
|
|
chr18_+_54682261
|
0.570
|
|
ZNF532
|
zinc finger protein 532
|
|
chr19_+_60487578
|
0.570
|
|
BRSK1
|
BR serine/threonine kinase 1
|
|
chr5_-_176857158
|
0.569
|
|
PDLIM7
|
PDZ and LIM domain 7 (enigma)
|
|
chr9_-_35101592
|
0.568
|
|
KIAA1539
|
KIAA1539
|
|
chr11_-_125730655
|
0.566
|
|
FLJ39051
|
hypothetical LOC399972
|
|
chr1_+_35031185
|
0.566
|
NM_002060
|
GJA4
|
gap junction protein, alpha 4, 37kDa
|
|
chr22_-_17546296
|
0.566
|
NM_005984
|
SLC25A1
|
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1
|
|
chr4_+_154606900
|
0.565
|
NM_001131007
NM_015196
|
KIAA0922
|
KIAA0922
|
|
chr2_-_120697148
|
0.565
|
|
TMEM185B
|
transmembrane protein 185B (pseudogene)
|
|
chr3_+_38182251
|
0.565
|
|
OXSR1
|
oxidative-stress responsive 1
|
|
chr22_+_41877470
|
0.564
|
NM_000714
NM_007311
|
TSPO
|
translocator protein (18kDa)
|