|
chr4_-_809879
|
3.501
|
NM_006651
|
CPLX1
|
complexin 1
|
|
chr7_+_5289079
|
2.699
|
NM_001040661
NM_153247
|
SLC29A4
|
solute carrier family 29 (nucleoside transporters), member 4
|
|
chr10_+_102495327
|
1.911
|
NM_000278
NM_003987
NM_003988
NM_003989
NM_003990
|
PAX2
|
paired box 2
|
|
chr16_+_29724926
|
1.901
|
|
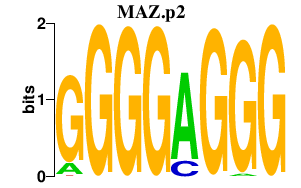
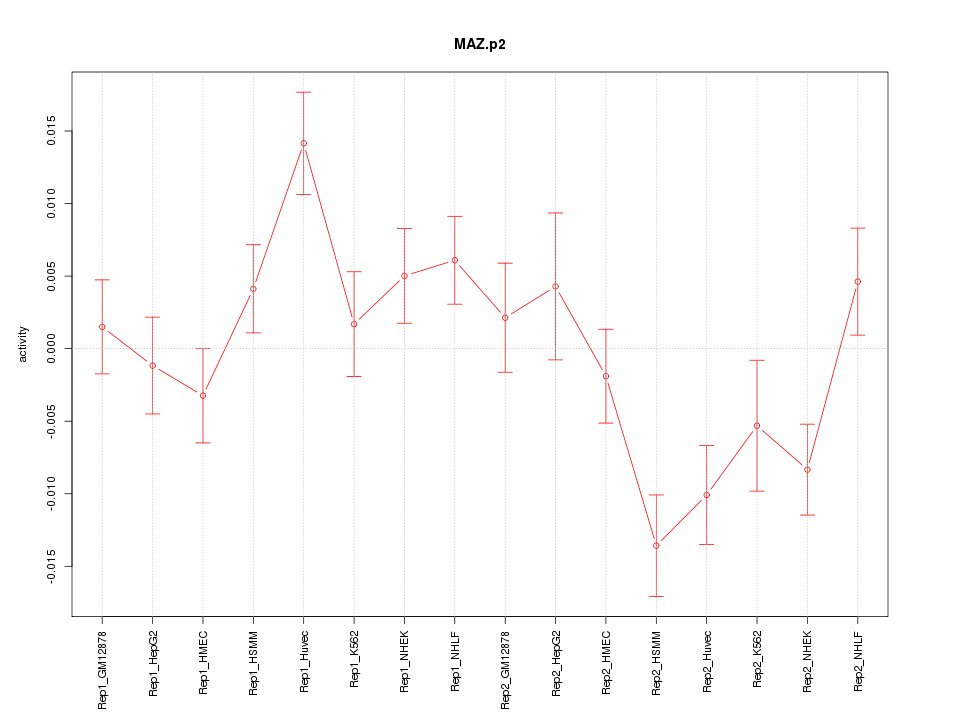
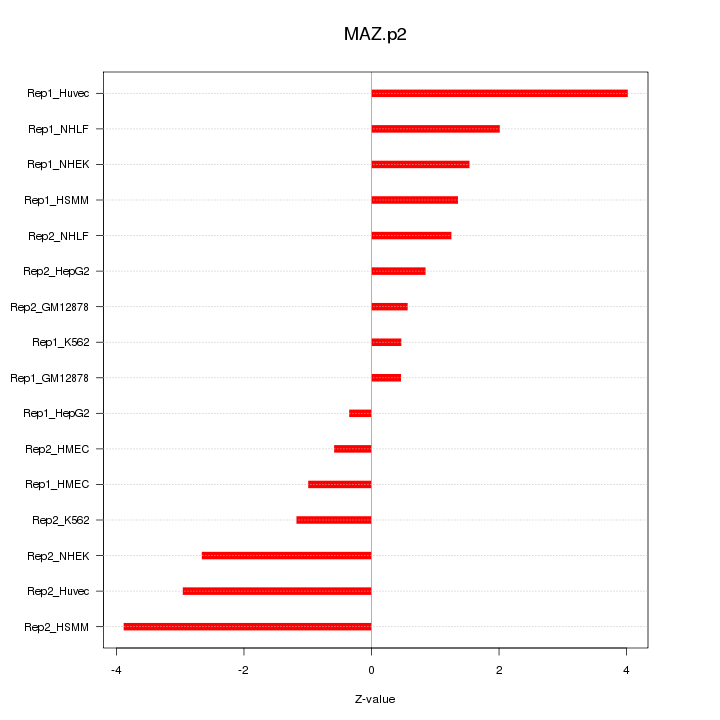
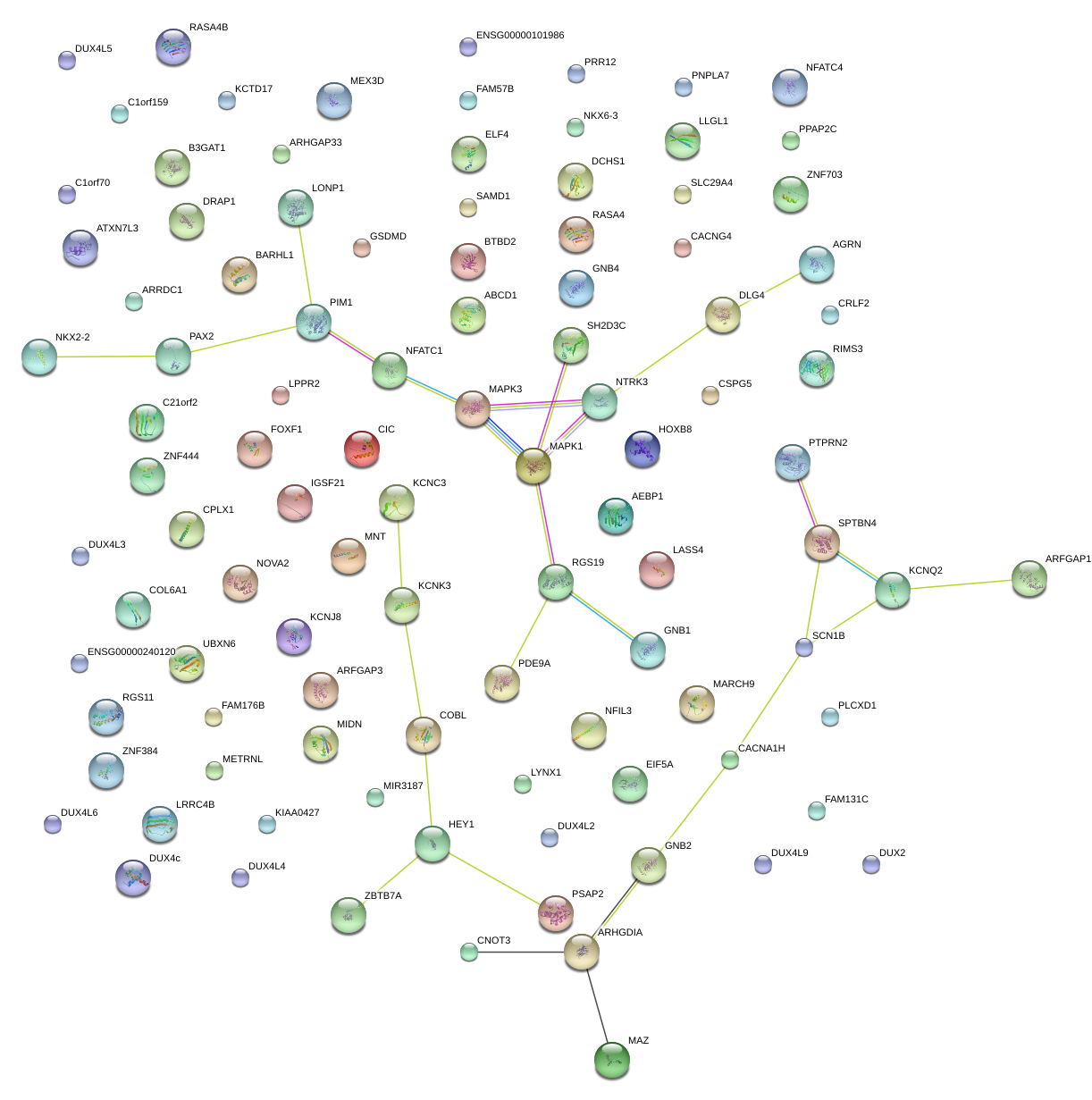
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor)
|
|
chr19_-_242168
|
1.767
|
NM_177543
|
PPAP2C
|
phosphatidic acid phosphatase type 2C
|
|
chrX_-_1291526
|
1.757
|
NM_001012288
NM_022148
|
CRLF2
|
cytokine receptor-like factor 2
|
|
chr19_-_242335
|
1.681
|
NM_003712
NM_177526
|
PPAP2C
|
phosphatidic acid phosphatase type 2C
|
|
chr21_+_46226072
|
1.662
|
NM_001848
|
COL6A1
|
collagen, type VI, alpha 1
|
|
chr17_-_39632845
|
1.660
|
|
ATXN7L3
|
ataxin 7-like 3
|
|
chr19_+_47480445
|
1.500
|
NM_015125
|
CIC
|
capicua homolog (Drosophila)
|
|
chr19_-_4016467
|
1.468
|
|
ZBTB7A
|
zinc finger and BTB domain containing 7A
|
|
chr1_-_36562341
|
1.401
|
NM_018166
|
FAM176B
|
family with sequence similarity 176, member B
|
|
chr19_+_54786723
|
1.375
|
NM_020719
|
PRR12
|
proline rich 12
|
|
chr19_-_1966628
|
1.356
|
NM_017797
|
BTBD2
|
BTB (POZ) domain containing 2
|
|
chr17_+_62391216
|
1.332
|
NM_014405
|
CACNG4
|
calcium channel, voltage-dependent, gamma subunit 4
|
|
chr9_+_139619969
|
1.310
|
|
ARRDC1
|
arrestin domain containing 1
|
|
chr18_+_75256954
|
1.308
|
|
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1
|
|
chr6_+_37245860
|
1.298
|
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr16_-_29949637
|
1.283
|
NM_031478
|
FAM57B
|
family with sequence similarity 57, member B
|
|
chr14_+_23907065
|
1.257
|
NM_001198965
NM_004554
|
NFATC4
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4
|
|
chr1_-_1465602
|
1.256
|
NM_001114748
|
C1orf70
|
chromosome 1 open reading frame 70
|
|
chr2_+_26768894
|
1.254
|
NM_002246
|
KCNK3
|
potassium channel, subfamily K, member 3
|
|
chr12_-_6668680
|
1.238
|
|
ZNF384
|
zinc finger protein 384
|
|
chr20_-_62181231
|
1.217
|
NM_005873
|
RGS19
|
regulator of G-protein signaling 19
|
|
chr21_-_44583505
|
1.207
|
|
C21orf2
|
chromosome 21 open reading frame 2
|
|
chr12_-_21818881
|
1.204
|
NM_004982
|
KCNJ8
|
potassium inwardly-rectifying channel, subfamily J, member 8
|
|
chr16_+_29725311
|
1.198
|
NM_001042539
NM_002383
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor)
|
|
chr19_+_61344501
|
1.190
|
|
ZNF444
|
zinc finger protein 444
|
|
chr7_-_51351825
|
1.183
|
|
COBL
|
cordon-bleu homolog (mouse)
|
|
chr12_+_56435042
|
1.165
|
NM_138396
|
MARCH9
|
membrane-associated ring finger (C3HC4) 9
|
|
chr19_+_11327061
|
1.158
|
NM_001170635
NM_022737
|
LPPR2
|
lipid phosphate phosphatase-related protein type 2
|
|
chr11_-_6633427
|
1.155
|
NM_003737
|
DCHS1
|
dachsous 1 (Drosophila)
|
|
chr8_-_41624030
|
1.133
|
NM_152568
|
NKX6-3
|
NK6 homeobox 3
|
|
chr19_-_14062230
|
1.129
|
NM_138352
|
SAMD1
|
sterile alpha motif domain containing 1
|
|
chr20_-_21442663
|
1.126
|
NM_002509
|
NKX2-2
|
NK2 homeobox 2
|
|
chrX_+_152643516
|
1.110
|
NM_000033
|
ABCD1
|
ATP-binding cassette, sub-family D (ALD), member 1
|
|
chr20_-_62181588
|
1.100
|
NM_001039467
|
RGS19
|
regulator of G-protein signaling 19
|
|
chr19_+_8180179
|
1.099
|
NM_024552
|
LASS4
|
LAG1 homolog, ceramide synthase 4
|
|
chr19_-_772913
|
1.093
|
NM_024888
|
LPPR3
|
lipid phosphate phosphatase-related protein type 3
|
|
chr4_+_191245840
|
1.081
|
NM_033178
NM_012147
NM_001177376
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L4
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 4
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr3_-_47595190
|
1.070
|
NM_006574
|
CSPG5
|
chondroitin sulfate proteoglycan 5 (neuroglycan C)
|
|
chr9_-_139564656
|
1.060
|
NM_001098537
NM_152286
|
PNPLA7
|
patatin-like phospholipase domain containing 7
|
|
chr17_+_7151984
|
1.053
|
NM_001143761
|
EIF5A
|
eukaryotic translation initiation factor 5A
|
|
chr11_+_65443303
|
1.046
|
NM_006442
|
DRAP1
|
DR1-associated protein 1 (negative cofactor 2 alpha)
|
|
chr19_+_61344341
|
1.036
|
NM_018337
|
ZNF444
|
zinc finger protein 444
|
|
chr22_+_49459935
|
1.036
|
NM_001080420
|
SHANK3
|
SH3 and multiple ankyrin repeat domains 3
|
|
chr11_-_133786935
|
1.035
|
NM_018644
NM_054025
|
B3GAT1
|
beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P)
|
|
chr18_+_44319365
|
1.032
|
NM_001142397
NM_014772
|
CTIF
|
CBP80/20-dependent translation initiation factor
|
|
chr1_-_40903897
|
1.030
|
NM_014747
|
RIMS3
|
regulating synaptic membrane exocytosis 3
|
|
chr2_+_100087812
|
1.029
|
|
|
|
|
chr19_-_55524445
|
1.027
|
NM_004977
|
KCNC3
|
potassium voltage-gated channel, Shaw-related subfamily, member 3
|
|
chr19_-_4408789
|
1.020
|
NM_025241
|
UBXN6
|
UBX domain protein 6
|
|
chr17_-_2250967
|
1.020
|
NM_020310
|
MNT
|
MAX binding protein
|
|
chr9_-_129557373
|
1.018
|
NM_001142534
NM_001142533
|
SH2D3C
|
SH2 domain containing 3C
|
|
chr8_-_143855438
|
1.002
|
NM_177476
NM_177477
|
LYNX1
|
Ly6/neurotoxin 1
|
|
chr12_-_6668787
|
0.997
|
NM_001039918
NM_133476
|
ZNF384
|
zinc finger protein 384
|
|
chr8_+_144706699
|
0.992
|
NM_001166237
|
GSDMD
|
gasdermin D
|
|
chr19_+_1200882
|
0.991
|
|
MIDN
|
midnolin
|
|
chr16_+_1143241
|
0.982
|
NM_001005407
NM_021098
|
CACNA1H
|
calcium channel, voltage-dependent, T type, alpha 1H subunit
|
|
chr19_+_11327173
|
0.981
|
|
LPPR2
|
lipid phosphate phosphatase-related protein type 2
|
|
chr17_-_7061641
|
0.978
|
NM_001128827
|
DLG4
|
discs, large homolog 4 (Drosophila)
|
|
chr16_-_265868
|
0.973
|
NM_003834
NM_183337
|
RGS11
|
regulator of G-protein signaling 11
|
|
chr17_+_7151653
|
0.972
|
|
EIF5A
|
eukaryotic translation initiation factor 5A
|
|
chr18_+_75256915
|
0.949
|
|
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1
|
|
chr19_-_772966
|
0.948
|
|
LPPR3
|
lipid phosphate phosphatase-related protein type 3
|
|
chr20_-_61574259
|
0.944
|
NM_004518
NM_172106
NM_172107
NM_172108
NM_172109
|
KCNQ2
|
potassium voltage-gated channel, KQT-like subfamily, member 2
|
|
chr9_+_139619957
|
0.941
|
|
ARRDC1
|
arrestin domain containing 1
|
|
chr1_+_35104120
|
0.937
|
|
|
|
|
chr21_+_42946719
|
0.934
|
NM_001001567
NM_001001568
NM_001001569
NM_001001570
NM_001001571
NM_001001572
NM_001001573
NM_001001574
NM_001001575
NM_001001576
NM_001001577
NM_001001578
NM_001001579
NM_001001580
NM_001001581
NM_001001582
NM_001001583
NM_001001584
NM_001001585
NM_002606
|
PDE9A
|
phosphodiesterase 9A
|
|
chr12_-_6668880
|
0.932
|
NM_001039917
NM_001039919
NM_001039920
NM_001135734
|
ZNF384
|
zinc finger protein 384
|
|
chr19_+_45728270
|
0.926
|
|
SPTBN4
|
spectrin, beta, non-erythrocytic 4
|
|
chrX_+_138060
|
0.924
|
NM_018390
|
PLCXD1
|
phosphatidylinositol-specific phospholipase C, X domain containing 1
|
|
chr9_+_134447795
|
0.923
|
NM_020064
|
BARHL1
|
BarH-like homeobox 1
|
|
chr7_+_44110527
|
0.915
|
|
AEBP1
|
AE binding protein 1
|
|
chr16_+_85101734
|
0.897
|
|
FOXF1
|
forkhead box F1
|
|
chr17_+_78630793
|
0.889
|
NM_001004431
|
METRNL
|
meteorin, glial cell differentiation regulator-like
|
|
chr19_+_45728207
|
0.880
|
NM_025213
|
SPTBN4
|
spectrin, beta, non-erythrocytic 4
|
|
chr19_+_59333232
|
0.878
|
NM_014516
|
CNOT3
|
CCR4-NOT transcription complex, subunit 3
|
|
chr17_-_77422463
|
0.875
|
|
ARHGDIA
|
Rho GDP dissociation inhibitor (GDI) alpha
|
|
chr19_+_40958206
|
0.869
|
NM_001172630
NM_052948
|
ARHGAP33
|
Rho GTPase activating protein 33
|
|
chr8_+_37672427
|
0.866
|
NM_025069
|
ZNF703
|
zinc finger protein 703
|
|
chr1_+_945472
|
0.865
|
|
AGRN
|
agrin
|
|
chr15_-_86600658
|
0.865
|
NM_001007156
NM_001012338
NM_002530
|
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3
|
|
chr7_-_158073121
|
0.856
|
NM_002847
NM_130842
NM_130843
|
PTPRN2
|
protein tyrosine phosphatase, receptor type, N polypeptide 2
|
|
chr19_-_51168396
|
0.854
|
NM_002516
|
NOVA2
|
neuro-oncological ventral antigen 2
|
|
chr19_-_55763091
|
0.852
|
NM_001080457
|
LRRC4B
|
leucine rich repeat containing 4B
|
|
chr7_-_102044374
|
0.851
|
|
RASA4
|
RAS p21 protein activator 4
|
|
chr22_+_35777561
|
0.850
|
NM_024681
|
KCTD17
|
potassium channel tetramerisation domain containing 17
|
|
chr7_+_100111689
|
0.841
|
|
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2
|
|
chr1_+_945331
|
0.834
|
NM_198576
|
AGRN
|
agrin
|
|
chr19_+_40213431
|
0.833
|
NM_001037
NM_199037
|
SCN1B
|
sodium channel, voltage-gated, type I, beta
|
|
chr18_+_75256702
|
0.833
|
NM_006162
NM_172388
NM_172390
|
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1
|
|
chr1_+_18306823
|
0.830
|
NM_032880
|
IGSF21
|
immunoglobin superfamily, member 21
|
|
chr1_-_16272523
|
0.825
|
NM_182623
|
FAM131C
|
family with sequence similarity 131, member C
|
|
chr16_-_30041767
|
0.823
|
|
MAPK3
|
mitogen-activated protein kinase 3
|
|
chr17_-_44047299
|
0.822
|
NM_024016
|
HOXB8
|
homeobox B8
|
|
chr8_-_80842452
|
0.816
|
NM_001040708
NM_012258
|
HEY1
|
hairy/enhancer-of-split related with YRPW motif 1
|
|
chr16_+_85101605
|
0.814
|
NM_001451
|
FOXF1
|
forkhead box F1
|
|
chr1_-_1041321
|
0.813
|
|
C1orf159
|
chromosome 1 open reading frame 159
|
|
chr19_-_1518875
|
0.811
|
NM_001174118
NM_203304
|
MEX3D
|
mex-3 homolog D (C. elegans)
|
|
chr11_+_65443452
|
0.810
|
|
DRAP1
|
DR1-associated protein 1 (negative cofactor 2 alpha)
|
|
chr20_+_61374628
|
0.808
|
|
ARFGAP1
|
ADP-ribosylation factor GTPase activating protein 1
|
|
chr19_+_540849
|
0.806
|
NM_001194
|
HCN2
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 2
|
|
chr16_-_70475381
|
0.803
|
|
ZNF821
|
zinc finger protein 821
|
|
chr7_+_100111712
|
0.800
|
|
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2
|
|
chr19_+_13924287
|
0.800
|
|
DCAF15
|
DDB1 and CUL4 associated factor 15
|
|
chr9_+_139619875
|
0.799
|
NM_152285
|
ARRDC1
|
arrestin domain containing 1
|
|
chr19_+_7874774
|
0.798
|
|
MAP2K7
|
mitogen-activated protein kinase kinase 7
|
|
chr20_+_62181878
|
0.794
|
NM_000913
NM_182647
|
OPRL1
|
opiate receptor-like 1
|
|
chr8_-_144750987
|
0.794
|
NM_001130053
NM_001130055
NM_001130056
NM_001195203
NM_001960
NM_032378
|
EEF1D
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein)
|
|
chr19_-_6230797
|
0.786
|
|
|
|
|
chr1_-_158335241
|
0.784
|
|
IGSF8
|
immunoglobulin superfamily, member 8
|
|
chr22_-_44015248
|
0.784
|
NM_001009880
|
KIAA0930
|
KIAA0930
|
|
chr9_+_139619977
|
0.782
|
|
ARRDC1
|
arrestin domain containing 1
|
|
chr11_-_2863536
|
0.779
|
NM_000076
NM_001122630
NM_001122631
|
CDKN1C
|
cyclin-dependent kinase inhibitor 1C (p57, Kip2)
|
|
chr4_+_191232653
|
0.777
|
NM_033178
NM_012147
NM_001177376
NM_001127387
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L4
DUX4L7
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 4
double homeobox 4 like 7
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr14_+_103621768
|
0.772
|
NM_001080464
|
ASPG
|
asparaginase homolog (S. cerevisiae)
|
|
chr1_+_154318954
|
0.771
|
|
LMNA
|
lamin A/C
|
|
chr20_-_30534848
|
0.766
|
NM_080616
|
C20orf112
|
chromosome 20 open reading frame 112
|
|
chr19_-_60783996
|
0.766
|
NM_152600
|
ZNF579
|
zinc finger protein 579
|
|
chr11_-_62145952
|
0.765
|
NM_012200
|
B3GAT3
|
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I)
|
|
chr14_-_22891865
|
0.763
|
NM_016609
|
SLC22A17
|
solute carrier family 22, member 17
|
|
chr2_-_241408296
|
0.760
|
NM_004321
|
KIF1A
|
kinesin family member 1A
|
|
chr11_-_62146144
|
0.760
|
|
B3GAT3
|
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I)
|
|
chr10_+_135343649
|
0.753
|
NM_012147
|
DUX2
|
double homeobox 2
|
|
chr20_+_34523231
|
0.753
|
NM_001042486
|
DLGAP4
|
discs, large (Drosophila) homolog-associated protein 4
|
|
chr11_-_69343128
|
0.750
|
NM_005247
|
FGF3
|
fibroblast growth factor 3
|
|
chr20_+_32610161
|
0.749
|
NM_032514
|
MAP1LC3A
|
microtubule-associated protein 1 light chain 3 alpha
|
|
chr11_-_61441536
|
0.746
|
NM_013401
|
RAB3IL1
|
RAB3A interacting protein (rabin3)-like 1
|
|
chr17_-_39556535
|
0.746
|
NM_001015053
NM_005474
|
HDAC5
|
histone deacetylase 5
|
|
chr19_-_1188840
|
0.746
|
NM_152769
|
C19orf26
|
chromosome 19 open reading frame 26
|
|
chr19_-_6230927
|
0.744
|
NM_005934
|
MLLT1
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 1
|
|
chr1_-_153429257
|
0.743
|
NM_001018016
NM_001018017
NM_001044390
NM_001044391
NM_001044392
NM_001044393
NM_002456
|
MUC1
|
mucin 1, cell surface associated
|
|
chr14_-_37134050
|
0.743
|
|
FOXA1
|
forkhead box A1
|
|
chr5_+_122452739
|
0.742
|
NM_001136239
|
PRDM6
|
PR domain containing 6
|
|
chr17_-_77422526
|
0.742
|
|
ARHGDIA
|
Rho GDP dissociation inhibitor (GDI) alpha
|
|
chr11_+_64608307
|
0.738
|
|
ZFPL1
|
zinc finger protein-like 1
|
|
chr19_-_1814425
|
0.732
|
|
KLF16
|
Kruppel-like factor 16
|
|
chr11_-_64247230
|
0.730
|
NM_015080
NM_138732
|
NRXN2
|
neurexin 2
|
|
chr12_+_50686990
|
0.729
|
NM_181711
|
GRASP
|
GRP1 (general receptor for phosphoinositides 1)-associated scaffold protein
|
|
chr16_-_2186465
|
0.729
|
NM_020764
|
CASKIN1
|
CASK interacting protein 1
|
|
chr11_+_63337607
|
0.728
|
|
C11orf84
|
chromosome 11 open reading frame 84
|
|
chr11_-_66897584
|
0.728
|
NM_013246
|
CLCF1
|
cardiotrophin-like cytokine factor 1
|
|
chr2_+_24152799
|
0.728
|
|
|
|
|
chr7_-_100595552
|
0.727
|
NM_003378
|
VGF
|
VGF nerve growth factor inducible
|
|
chr7_+_28415495
|
0.726
|
|
LOC401317
|
hypothetical LOC401317
|
|
chr6_+_1335067
|
0.724
|
NM_001452
|
FOXF2
|
forkhead box F2
|
|
chr15_+_88345612
|
0.723
|
NM_198526
|
ZNF710
|
zinc finger protein 710
|
|
chr19_-_52667056
|
0.723
|
NM_015063
|
SLC8A2
|
solute carrier family 8 (sodium/calcium exchanger), member 2
|
|
chr12_-_49897743
|
0.723
|
|
POU6F1
|
POU class 6 homeobox 1
|
|
chr10_+_101282679
|
0.722
|
NM_145285
|
NKX2-3
|
NK2 transcription factor related, locus 3 (Drosophila)
|
|
chr1_+_945399
|
0.722
|
|
|
|
|
chr14_-_20636337
|
0.719
|
|
ZNF219
|
zinc finger protein 219
|
|
chr19_+_1226510
|
0.712
|
NM_017914
|
C19orf24
|
chromosome 19 open reading frame 24
|
|
chr16_+_3036682
|
0.712
|
NM_022468
|
MMP25
|
matrix metallopeptidase 25
|
|
chr11_+_279113
|
0.711
|
NM_025092
|
ATHL1
|
ATH1, acid trehalase-like 1 (yeast)
|
|
chr22_-_29052863
|
0.710
|
NM_031937
|
TBC1D10A
|
TBC1 domain family, member 10A
|
|
chr1_+_153241665
|
0.707
|
NM_015872
|
ZBTB7B
|
zinc finger and BTB domain containing 7B
|
|
chr19_+_60803412
|
0.707
|
NM_153219
|
ZNF524
|
zinc finger protein 524
|
|
chr15_+_72209795
|
0.705
|
NM_020851
|
ISLR2
|
immunoglobulin superfamily containing leucine-rich repeat 2
|
|
chr19_+_13924318
|
0.705
|
NM_138353
|
DCAF15
|
DDB1 and CUL4 associated factor 15
|
|
chr4_+_3264509
|
0.705
|
|
RGS12
|
regulator of G-protein signaling 12
|
|
chr7_-_149101227
|
0.704
|
NM_207336
|
ZNF467
|
zinc finger protein 467
|
|
chr5_-_179166378
|
0.704
|
NM_014275
|
MGAT4B
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme B
|
|
chr1_-_27802688
|
0.703
|
NM_001029882
|
AHDC1
|
AT hook, DNA binding motif, containing 1
|
|
chr22_-_19122047
|
0.702
|
|
SCARF2
|
scavenger receptor class F, member 2
|
|
chr9_+_131467837
|
0.701
|
|
PRRX2
|
paired related homeobox 2
|
|
chr12_+_93066370
|
0.700
|
NM_005761
|
PLXNC1
|
plexin C1
|
|
chr20_+_34523555
|
0.699
|
NM_183006
|
DLGAP4
|
discs, large (Drosophila) homolog-associated protein 4
|
|
chr8_-_144722141
|
0.697
|
|
C8orf73
|
chromosome 8 open reading frame 73
|
|
chr22_-_19122107
|
0.697
|
NM_153334
NM_182895
|
SCARF2
|
scavenger receptor class F, member 2
|
|
chr12_+_47658502
|
0.695
|
NM_005430
|
WNT1
|
wingless-type MMTV integration site family, member 1
|
|
chr17_-_39556483
|
0.695
|
|
HDAC5
|
histone deacetylase 5
|
|
chr5_+_139155569
|
0.692
|
NM_032289
|
PSD2
|
pleckstrin and Sec7 domain containing 2
|
|
chr15_-_38953628
|
0.691
|
NM_133639
|
RHOV
|
ras homolog gene family, member V
|
|
chr19_+_748390
|
0.688
|
NM_002819
NM_031990
NM_031991
NM_175847
|
PTBP1
|
polypyrimidine tract binding protein 1
|
|
chr17_+_58954418
|
0.686
|
NM_030779
NM_173092
|
KCNH6
|
potassium voltage-gated channel, subfamily H (eag-related), member 6
|
|
chr12_+_55896844
|
0.685
|
NM_007224
|
NXPH4
|
neurexophilin 4
|
|
chr4_-_1232742
|
0.684
|
|
CTBP1
|
C-terminal binding protein 1
|
|
chr14_-_20636568
|
0.682
|
NM_016423
|
ZNF219
|
zinc finger protein 219
|
|
chr19_+_52451642
|
0.682
|
|
CCDC9
|
coiled-coil domain containing 9
|
|
chr12_-_47651812
|
0.680
|
NM_003394
|
WNT10B
|
wingless-type MMTV integration site family, member 10B
|
|
chr18_+_75256931
|
0.680
|
|
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1
|
|
chr5_-_1165106
|
0.680
|
NM_006598
|
SLC12A7
|
solute carrier family 12 (potassium/chloride transporters), member 7
|
|
chr5_+_109052984
|
0.680
|
|
|
|
|
chr12_-_53268538
|
0.679
|
NM_006741
|
PPP1R1A
|
protein phosphatase 1, regulatory (inhibitor) subunit 1A
|
|
chr20_-_604822
|
0.676
|
NM_033129
|
SCRT2
|
scratch homolog 2, zinc finger protein (Drosophila)
|
|
chr1_-_15723363
|
0.674
|
NM_001229
NM_032996
|
CASP9
|
caspase 9, apoptosis-related cysteine peptidase
|
|
chr16_+_19087035
|
0.672
|
NM_016524
|
SYT17
|
synaptotagmin XVII
|
|
chr19_-_16998449
|
0.671
|
NM_015692
|
CPAMD8
|
C3 and PZP-like, alpha-2-macroglobulin domain containing 8
|
|
chr9_+_139619917
|
0.671
|
|
ARRDC1
|
arrestin domain containing 1
|
|
chr12_+_6800950
|
0.670
|
NM_014449
NM_019858
|
GPR162
|
G protein-coupled receptor 162
|
|
chr3_-_52419086
|
0.670
|
|
BAP1
|
BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase)
|
|
chr19_+_45996881
|
0.669
|
NM_080732
|
EGLN2
|
egl nine homolog 2 (C. elegans)
|
|
chr19_+_39664706
|
0.668
|
NM_001080436
|
WTIP
|
Wilms tumor 1 interacting protein
|
|
chr17_-_77602938
|
0.667
|
NM_002917
|
RFNG
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr19_-_11477395
|
0.666
|
|
ZNF653
|
zinc finger protein 653
|
|
chr11_+_65049152
|
0.666
|
|
SCYL1
|
SCY1-like 1 (S. cerevisiae)
|
|
chr1_-_175400441
|
0.665
|
|
ASTN1
|
astrotactin 1
|
|
chr2_+_936534
|
0.665
|
NM_018968
|
SNTG2
|
syntrophin, gamma 2
|