|
chr1_+_65503011
|
1.306
|
NM_014787
|
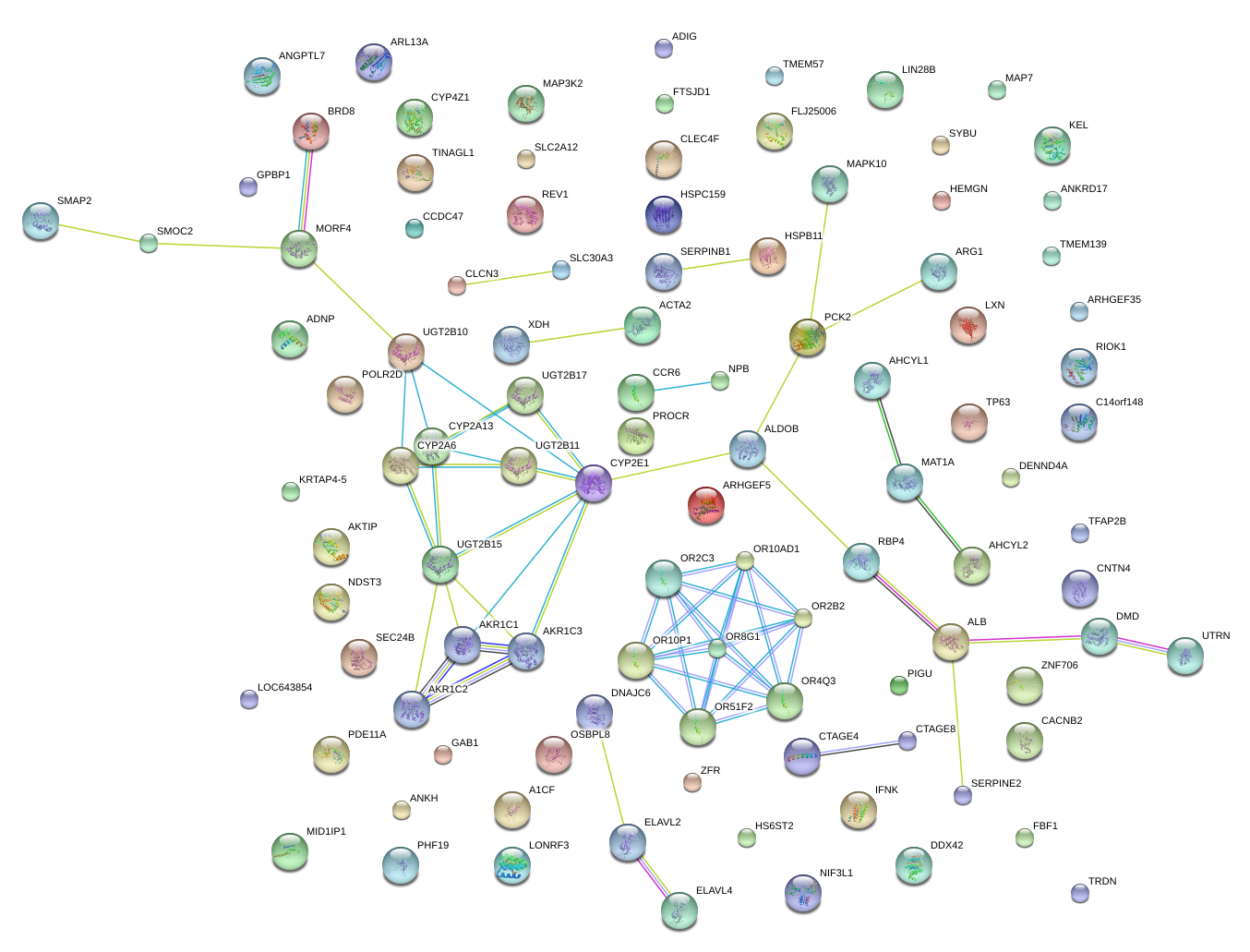
DNAJC6
|
DnaJ (Hsp40) homolog, subfamily C, member 6
|
|
chr6_+_167445284
|
1.107
|
NM_004367
|
CCR6
|
chemokine (C-C motif) receptor 6
|
|
chr11_-_10487212
|
1.057
|
NM_001190702
|
MTRNR2L8
|
MT-RNR2-like 8
|
|
chrX_+_117992603
|
1.027
|
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3
|
|
chr7_-_142369519
|
0.941
|
|
KEL
|
Kell blood group, metallo-endopeptidase
|
|
chr7_-_142369534
|
0.811
|
NM_000420
|
KEL
|
Kell blood group, metallo-endopeptidase
|
|
chr10_-_82039158
|
0.794
|
NM_000429
|
MAT1A
|
methionine adenosyltransferase I, alpha
|
|
chr2_+_201464884
|
0.757
|
NM_001142356
|
NIF3L1
|
NIF3 NGG1 interacting factor 3-like 1 (S. pombe)
|
|
chr10_-_52315436
|
0.743
|
NM_001198818
NM_001198819
NM_001198820
NM_014576
NM_138932
NM_138933
|
A1CF
|
APOBEC1 complementation factor
|
|
chr14_+_23633101
|
0.722
|
NM_001018073
NM_004563
|
PCK2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial)
|
|
chr6_-_123999640
|
0.679
|
NM_006073
|
TRDN
|
triadin
|
|
chr5_-_14924555
|
0.657
|
NM_054027
|
ANKH
|
ankylosis, progressive homolog (mouse)
|
|
chr2_-_99472843
|
0.654
|
|
REV1
|
REV1 homolog (S. cerevisiae)
|
|
chr9_+_27514311
|
0.634
|
NM_020124
|
IFNK
|
interferon, kappa
|
|
chr4_+_110574376
|
0.601
|
NM_001042734
NM_006323
|
SEC24B
|
SEC24 family, member B (S. cerevisiae)
|
|
chr9_-_122679387
|
0.597
|
NM_001009936
NM_015651
|
PHF19
|
PHD finger protein 19
|
|
chr6_-_27988152
|
0.582
|
NM_033057
|
OR2B2
|
olfactory receptor, family 2, subfamily B, member 2
|
|
chr1_+_11171967
|
0.573
|
NM_021146
|
ANGPTL7
|
angiopoietin-like 7
|
|
chr9_-_122679283
|
0.571
|
|
PHF19
|
PHD finger protein 19
|
|
chr4_+_170817787
|
0.557
|
|
CLCN3
|
chloride channel 3
|
|
chrX_+_100111352
|
0.555
|
NM_001012990
NM_001162490
NM_001162491
|
ARL13A
|
ADP-ribosylation factor-like 13A
|
|
chr6_-_136889302
|
0.552
|
|
MAP7
|
microtubule-associated protein 7
|
|
chr12_+_54316910
|
0.547
|
NM_206899
|
OR10P1
|
olfactory receptor, family 10, subfamily P, member 1
|
|
chr1_+_25629968
|
0.538
|
NM_018202
|
TMEM57
|
transmembrane protein 57
|
|
chr3_+_2528272
|
0.536
|
|
CNTN4
|
contactin 4
|
|
chr9_+_126460511
|
0.536
|
|
|
|
|
chr8_-_102287083
|
0.528
|
NM_001042510
NM_016096
|
ZNF706
|
zinc finger protein 706
|
|
chr16_-_69876262
|
0.526
|
|
FTSJD1
|
FtsJ methyltransferase domain containing 1
|
|
chr2_-_63518589
|
0.517
|
NM_001042692
|
WDPCP
|
WD repeat containing planar cell polarity effector
|
|
chr17_+_59205289
|
0.516
|
NM_007372
NM_203499
|
DDX42
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 42
|
|
chr10_+_135190855
|
0.511
|
NM_000773
|
CYP2E1
|
cytochrome P450, family 2, subfamily E, polypeptide 1
|
|
chr4_+_110574420
|
0.508
|
|
SEC24B
|
SEC24 family, member B (S. cerevisiae)
|
|
chr15_-_63871492
|
0.502
|
|
DENND4A
|
DENN/MADD domain containing 4A
|
|
chr7_+_142692165
|
0.496
|
NM_153345
|
TMEM139
|
transmembrane protein 139
|
|
chr12_-_46883341
|
0.490
|
NM_001004134
|
OR10AD1
|
olfactory receptor, family 10, subfamily AD, member 1
|
|
chr1_+_50348052
|
0.488
|
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr17_-_23965305
|
0.484
|
NM_001174103
|
SGK494
|
uncharacterized serine/threonine-protein kinase SgK494
|
|
chr4_+_74488910
|
0.482
|
|
ALB
|
albumin
|
|
chr8_-_110730183
|
0.478
|
NM_001099747
NM_001099748
NM_001099749
NM_001099750
NM_017786
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr1_+_47305633
|
0.472
|
NM_178134
|
CYP4Z1
|
cytochrome P450, family 4, subfamily Z, polypeptide 1
|
|
chr5_-_79982584
|
0.472
|
NM_001190470
|
MTRNR2L2
|
MT-RNR2-like 2
|
|
chr6_+_50894397
|
0.465
|
NM_003221
|
TFAP2B
|
transcription factor AP-2 beta (activating enhancer binding protein 2 beta)
|
|
chr17_-_59204622
|
0.464
|
|
CCDC47
|
coiled-coil domain containing 47
|
|
chr6_-_2785682
|
0.455
|
|
SERPINB1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1
|
|
chr17_+_59205244
|
0.453
|
|
|
|
|
chr2_-_99472910
|
0.453
|
NM_001037872
NM_016316
|
REV1
|
REV1 homolog (S. cerevisiae)
|
|
chr4_+_74488947
|
0.453
|
|
ALB
|
albumin
|
|
chr7_+_143511480
|
0.448
|
NM_198495
|
CTAGE4
CTAGE8
|
CTAGE family, member 4
CTAGE family, member 8
|
|
chr2_-_178461710
|
0.443
|
NM_001077196
|
PDE11A
|
phosphodiesterase 11A
|
|
chr20_-_48980858
|
0.443
|
NM_015339
NM_181442
|
ADNP
|
activity-dependent neuroprotector homeobox
|
|
chrX_-_131919896
|
0.414
|
|
HS6ST2
|
heparan sulfate 6-O-sulfotransferase 2
|
|
chr1_+_40611955
|
0.411
|
NM_022733
|
SMAP2
|
small ArfGAP2
|
|
chr8_-_102287044
|
0.405
|
NM_001042511
|
ZNF706
|
zinc finger protein 706
|
|
chr20_+_33223434
|
0.405
|
NM_006404
|
PROCR
|
protein C receptor, endothelial
|
|
chr4_-_74307637
|
0.402
|
|
ANKRD17
|
ankyrin repeat domain 17
|
|
chr17_-_36559579
|
0.401
|
NM_033188
|
KRTAP4-5
|
keratin associated protein 4-5
|
|
chr6_+_105511615
|
0.400
|
NM_001004317
|
LIN28B
|
lin-28 homolog B (C. elegans)
|
|
chr4_+_144477432
|
0.399
|
NM_002039
NM_207123
|
GAB1
|
GRB2-associated binding protein 1
|
|
chr6_-_136888771
|
0.398
|
NM_001198608
NM_001198609
NM_001198611
|
MAP7
|
microtubule-associated protein 7
|
|
chr5_+_56507029
|
0.392
|
NM_001127235
|
GPBP1
|
GC-rich promoter binding protein 1
|
|
chr2_-_70901186
|
0.392
|
NM_173535
|
CLEC4F
|
C-type lectin domain family 4, member F
|
|
chr11_+_4799126
|
0.388
|
NM_001004753
|
OR51F2
|
olfactory receptor, family 51, subfamily F, member 2
|
|
chr10_+_18589589
|
0.380
|
NM_000724
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit
|
|
chr6_-_134415311
|
0.375
|
NM_145176
|
SLC2A12
|
solute carrier family 2 (facilitated glucose transporter), member 12
|
|
chr4_+_119174947
|
0.373
|
NM_004784
|
NDST3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3
|
|
chr1_-_54183833
|
0.370
|
NM_016126
|
HSPB11
|
heat shock protein family B (small), member 11
|
|
chrX_+_117992623
|
0.368
|
NM_001031855
NM_024778
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3
|
|
chr4_-_174774368
|
0.367
|
NM_006792
|
MORF4
|
mortality factor 4
|
|
chr14_-_76958984
|
0.366
|
NM_138791
NM_001113475
|
C14orf148
|
chromosome 14 open reading frame 148
|
|
chr17_+_55269974
|
0.361
|
|
|
|
|
chr5_-_32424444
|
0.353
|
|
ZFR
|
zinc finger RNA binding protein
|
|
chr11_+_123639909
|
0.348
|
NM_001005198
|
OR8G5
|
olfactory receptor, family 8, subfamily G, member 5
|
|
chr16_-_52094613
|
0.346
|
NM_001012398
NM_022476
|
AKTIP
|
AKT interacting protein
|
|
chr2_-_31491057
|
0.346
|
|
XDH
|
xanthine dehydrogenase
|
|
chr19_-_46080496
|
0.343
|
NM_000764
NM_030589
|
CYP2A7
|
cytochrome P450, family 2, subfamily A, polypeptide 7
|
|
chr1_-_245763763
|
0.342
|
NM_198074
|
OR2C3
|
olfactory receptor, family 2, subfamily C, member 3
|
|
chr2_+_64540843
|
0.338
|
|
HSPC159
|
galectin-related protein
|
|
chrX_+_38548316
|
0.337
|
|
MID1IP1
|
MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish))
|
|
chr20_+_36643251
|
0.335
|
NM_001018082
|
ADIG
|
adipogenin
|
|
chr6_+_7347878
|
0.332
|
NM_153005
|
RIOK1
|
RIO kinase 1 (yeast)
|
|
chr6_+_131936057
|
0.332
|
NM_000045
|
ARG1
|
arginase, liver
|
|
chrX_-_33139349
|
0.329
|
NM_004006
|
DMD
|
dystrophin
|
|
chr2_-_224611568
|
0.329
|
NM_001136530
|
SERPINE2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2
|
|
chr10_-_95350949
|
0.326
|
NM_006744
|
RBP4
|
retinol binding protein 4, plasma
|
|
chr4_-_69570919
|
0.321
|
NM_001076
|
UGT2B15
|
UDP glucuronosyltransferase 2 family, polypeptide B15
|
|
chr3_-_159872960
|
0.316
|
NM_020169
|
LXN
|
latexin
|
|
chr3_+_190831909
|
0.312
|
NM_001114978
NM_001114979
NM_003722
|
TP63
|
tumor protein p63
|
|
chr1_+_40612016
|
0.309
|
|
SMAP2
|
small ArfGAP2
|
|
chr7_+_128802719
|
0.309
|
NM_001130722
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr14_+_19285426
|
0.309
|
NM_172194
|
OR4Q3
|
olfactory receptor, family 4, subfamily Q, member 3
|
|
chr2_-_127817143
|
0.308
|
NM_006609
|
MAP3K2
|
mitogen-activated protein kinase kinase kinase 2
|
|
chr9_-_99746923
|
0.308
|
NM_018437
|
HEMGN
|
hemogen
|
|
chr9_-_103237851
|
0.307
|
NM_000035
|
ALDOB
|
aldolase B, fructose-bisphosphate
|
|
chr10_+_4995597
|
0.307
|
|
AKR1C1
AKR1C3
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr17_-_70901303
|
0.306
|
|
GRB2
|
growth factor receptor-bound protein 2
|
|
chrX_-_131375217
|
0.305
|
NM_001170701
NM_001170702
NM_001170703
|
MBNL3
|
muscleblind-like 3 (Drosophila)
|
|
chrX_+_41191630
|
0.304
|
NM_022567
|
NYX
|
nyctalopin
|
|
chrX_+_38548394
|
0.303
|
|
MID1IP1
|
MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish))
|
|
chr12_-_44407515
|
0.302
|
|
LOC400027
|
hypothetical LOC400027
|
|
chr7_-_143597313
|
0.302
|
NM_198495
|
CTAGE4
CTAGE8
|
CTAGE family, member 4
CTAGE family, member 8
|
|
chr2_-_31491102
|
0.298
|
NM_000379
|
XDH
|
xanthine dehydrogenase
|
|
chr1_+_210805319
|
0.297
|
NM_001030287
|
ATF3
|
activating transcription factor 3
|
|
chr18_+_30875297
|
0.296
|
NM_014268
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2
|
|
chr12_-_44407756
|
0.295
|
|
LOC400027
|
hypothetical LOC400027
|
|
chr13_+_96672705
|
0.294
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr4_-_153493133
|
0.288
|
NM_018315
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr4_+_88187215
|
0.283
|
|
AFF1
|
AF4/FMR2 family, member 1
|
|
chr16_+_74868676
|
0.280
|
NM_033401
|
CNTNAP4
|
contactin associated protein-like 4
|
|
chr2_-_86276102
|
0.278
|
|
IMMT
|
inner membrane protein, mitochondrial
|
|
chr17_-_60439764
|
0.276
|
|
GNA13
|
guanine nucleotide binding protein (G protein), alpha 13
|
|
chr16_-_54424517
|
0.276
|
NM_001025194
NM_001025195
NM_001266
|
CES1
|
carboxylesterase 1
|
|
chr8_-_102286695
|
0.276
|
|
ZNF706
|
zinc finger protein 706
|
|
chr22_-_15682555
|
0.275
|
NM_175878
|
XKR3
|
XK, Kell blood group complex subunit-related family, member 3
|
|
chr6_+_101953389
|
0.275
|
|
GRIK2
|
glutamate receptor, ionotropic, kainate 2
|
|
chr7_-_141048358
|
0.270
|
NM_001080392
|
KIAA1147
|
KIAA1147
|
|
chrX_+_38548373
|
0.267
|
|
MID1IP1
|
MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish))
|
|
chr11_+_9552809
|
0.267
|
NM_001143976
|
WEE1
|
WEE1 homolog (S. pombe)
|
|
chr5_-_43593277
|
0.265
|
NM_183323
|
PAIP1
|
poly(A) binding protein interacting protein 1
|
|
chrX_+_38548016
|
0.265
|
NM_021242
|
MID1IP1
|
MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish))
|
|
chr17_-_26665144
|
0.265
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr1_+_205328834
|
0.264
|
NM_001017365
NM_001017366
|
C4BPB
|
complement component 4 binding protein, beta
|
|
chr17_-_59204685
|
0.264
|
NM_020198
|
CCDC47
|
coiled-coil domain containing 47
|
|
chr22_-_27787793
|
0.261
|
NM_015370
|
C22orf31
|
chromosome 22 open reading frame 31
|
|
chr2_-_38379031
|
0.260
|
|
ATL2
|
atlastin GTPase 2
|
|
chr12_-_116283793
|
0.260
|
NM_000620
|
NOS1
|
nitric oxide synthase 1 (neuronal)
|
|
chr8_-_86441115
|
0.258
|
NM_001164830
|
CA1
|
carbonic anhydrase I
|
|
chr22_-_48865980
|
0.257
|
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1
|
|
chr10_-_64698831
|
0.254
|
NM_004241
|
JMJD1C
|
jumonji domain containing 1C
|
|
chr12_-_68291208
|
0.253
|
NM_201550
|
LRRC10
|
leucine rich repeat containing 10
|
|
chr7_-_112515068
|
0.252
|
NM_001146265
NM_018970
|
GPR85
|
G protein-coupled receptor 85
|
|
chr3_+_134976965
|
0.252
|
|
TF
|
transferrin
|
|
chr4_-_153820535
|
0.252
|
NM_152680
|
TMEM154
|
transmembrane protein 154
|
|
chr10_+_68355797
|
0.251
|
NM_178011
|
LRRTM3
|
leucine rich repeat transmembrane neuronal 3
|
|
chr1_+_195213289
|
0.250
|
NM_030787
|
CFHR5
|
complement factor H-related 5
|
|
chr10_+_96512447
|
0.247
|
NM_000769
|
CYP2C19
|
cytochrome P450, family 2, subfamily C, polypeptide 19
|
|
chr1_+_149165438
|
0.246
|
NM_001145415
NM_012432
|
SETDB1
|
SET domain, bifurcated 1
|
|
chr11_+_93394025
|
0.245
|
NM_001098672
|
HEPHL1
|
hephaestin-like 1
|
|
chr7_-_143085704
|
0.245
|
NM_178561
|
CTAGE6P
|
CTAGE family, member 6, pseudogene
|
|
chr1_+_7767300
|
0.244
|
NM_016831
|
PER3
|
period homolog 3 (Drosophila)
|
|
chr2_-_74606819
|
0.244
|
NM_133637
|
DQX1
|
DEAQ box RNA-dependent ATPase 1
|
|
chr2_+_113542017
|
0.244
|
NM_173161
|
IL1F10
|
interleukin 1 family, member 10 (theta)
|
|
chr6_-_28429950
|
0.243
|
NM_145909
|
ZNF323
|
zinc finger protein 323
|
|
chr15_+_91244563
|
0.241
|
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr2_+_87536174
|
0.236
|
|
NCRNA00152
|
non-protein coding RNA 152
|
|
chr8_-_17797127
|
0.236
|
NM_147203
NM_201553
NM_004467
NM_201552
|
FGL1
|
fibrinogen-like 1
|
|
chr2_-_86276015
|
0.236
|
|
IMMT
|
inner membrane protein, mitochondrial
|
|
chr7_-_31347032
|
0.232
|
NM_022728
|
NEUROD6
|
neurogenic differentiation 6
|
|
chr17_-_36074812
|
0.230
|
NM_152349
|
KRT222
|
keratin 222
|
|
chr15_-_53996597
|
0.229
|
NM_198400
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr10_-_104201234
|
0.227
|
NM_024886
|
C10orf95
|
chromosome 10 open reading frame 95
|
|
chr13_+_96672541
|
0.227
|
NM_144778
NM_207304
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr1_+_61697209
|
0.226
|
|
NFIA
|
nuclear factor I/A
|
|
chr17_-_38979178
|
0.221
|
NM_001079675
|
ETV4
|
ets variant 4
|
|
chr19_-_53841064
|
0.219
|
NM_001217
|
CA11
|
carbonic anhydrase XI
|
|
chr6_-_24597717
|
0.218
|
NM_177483
NM_001503
|
GPLD1
|
glycosylphosphatidylinositol specific phospholipase D1
|
|
chr3_+_187813538
|
0.217
|
NM_001622
|
AHSG
|
alpha-2-HS-glycoprotein
|
|
chr2_-_202811459
|
0.214
|
NM_001005781
NM_001005782
NM_003352
|
SUMO1
|
SMT3 suppressor of mif two 3 homolog 1 (S. cerevisiae)
|
|
chrX_-_118583346
|
0.212
|
NM_001170569
|
CXorf56
|
chromosome X open reading frame 56
|
|
chr2_+_132196533
|
0.211
|
NM_013310
|
C2orf27A
|
chromosome 2 open reading frame 27A
|
|
chr12_+_109956210
|
0.211
|
NM_015267
|
CUX2
|
cut-like homeobox 2
|
|
chr2_+_161701711
|
0.208
|
NM_004180
NM_133484
|
TANK
|
TRAF family member-associated NFKB activator
|
|
chr6_+_158654890
|
0.208
|
|
|
|
|
chr12_-_73889777
|
0.207
|
NM_139136
NM_139137
NM_153748
|
KCNC2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2
|
|
chr6_-_132073849
|
0.205
|
NM_001145659
|
CTAGE9
|
CTAGE family, member 9
|
|
chr7_+_80105907
|
0.204
|
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr5_-_137876416
|
0.204
|
|
ETF1
|
eukaryotic translation termination factor 1
|
|
chr18_+_19286784
|
0.202
|
NM_003831
|
RIOK3
|
RIO kinase 3 (yeast)
|
|
chr14_+_101345878
|
0.199
|
NM_002719
NM_178586
NM_178587
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma
|
|
chr9_+_71009746
|
0.199
|
NM_001170415
NM_001170416
|
TJP2
|
tight junction protein 2 (zona occludens 2)
|
|
chr1_-_6218554
|
0.193
|
|
ICMT
|
isoprenylcysteine carboxyl methyltransferase
|
|
chr4_-_176970895
|
0.192
|
|
GPM6A
|
glycoprotein M6A
|
|
chr17_-_38978772
|
0.192
|
NM_001986
|
ETV4
|
ets variant 4
|
|
chr7_-_112515035
|
0.191
|
|
GPR85
|
G protein-coupled receptor 85
|
|
chr3_-_195670256
|
0.191
|
NM_024524
|
ATP13A3
|
ATPase type 13A3
|
|
chr19_-_40684638
|
0.190
|
NM_001035516
|
DMKN
|
dermokine
|
|
chr1_+_151081953
|
0.190
|
NM_001128600
|
LCE6A
|
late cornified envelope 6A
|
|
chr21_-_30836051
|
0.188
|
NM_181612
|
KRTAP19-6
|
keratin associated protein 19-6
|
|
chrX_-_84521052
|
0.187
|
|
POF1B
|
premature ovarian failure, 1B
|
|
chrX_-_118583401
|
0.186
|
NM_001170570
NM_022101
|
CXorf56
|
chromosome X open reading frame 56
|
|
chr1_-_6218576
|
0.185
|
|
ICMT
|
isoprenylcysteine carboxyl methyltransferase
|
|
chr4_+_74488835
|
0.185
|
NM_000477
|
ALB
|
albumin
|
|
chr21_-_14840506
|
0.184
|
NM_022136
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1
|
|
chr19_+_56671649
|
0.184
|
NM_001080405
|
CEACAM18
|
carcinoembryonic antigen-related cell adhesion molecule 18
|
|
chr21_+_37714470
|
0.183
|
NM_130438
NM_001396
|
DYRK1A
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A
|
|
chr19_-_6410735
|
0.180
|
NM_024103
|
SLC25A23
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23
|
|
chr16_+_76613912
|
0.180
|
NM_005752
|
CLEC3A
|
C-type lectin domain family 3, member A
|
|
chr9_+_96561750
|
0.180
|
NM_001193329
|
C9orf3
|
chromosome 9 open reading frame 3
|
|
chr1_-_150599105
|
0.179
|
NM_001014342
|
FLG2
|
filaggrin family member 2
|
|
chr10_-_69267774
|
0.177
|
NM_021800
NM_201262
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12
|
|
chr6_-_133076873
|
0.177
|
NM_004666
|
VNN1
|
vanin 1
|
|
chr12_-_28016182
|
0.177
|
NM_002820
NM_198965
|
PTHLH
|
parathyroid hormone-like hormone
|
|
chr8_-_91066060
|
0.175
|
NM_002485
|
NBN
|
nibrin
|
|
chr2_-_88208650
|
0.175
|
NM_001443
|
FABP1
|
fatty acid binding protein 1, liver
|
|
chr22_+_38627031
|
0.172
|
NM_004810
|
GRAP2
|
GRB2-related adaptor protein 2
|
|
chr13_-_99983945
|
0.170
|
NM_033110
|
A2LD1
|
AIG2-like domain 1
|
|
chr2_-_86276403
|
0.169
|
NM_001100169
NM_001100170
NM_006839
|
IMMT
|
inner membrane protein, mitochondrial
|
|
chr11_+_65026382
|
0.168
|
|
|
|
|
chr10_-_69125932
|
0.167
|
NM_013266
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3
|
|
chr22_+_20252020
|
0.167
|
|
UBE2L3
|
ubiquitin-conjugating enzyme E2L 3
|
|
chr16_-_54424473
|
0.166
|
|
CES1
|
carboxylesterase 1
|