|
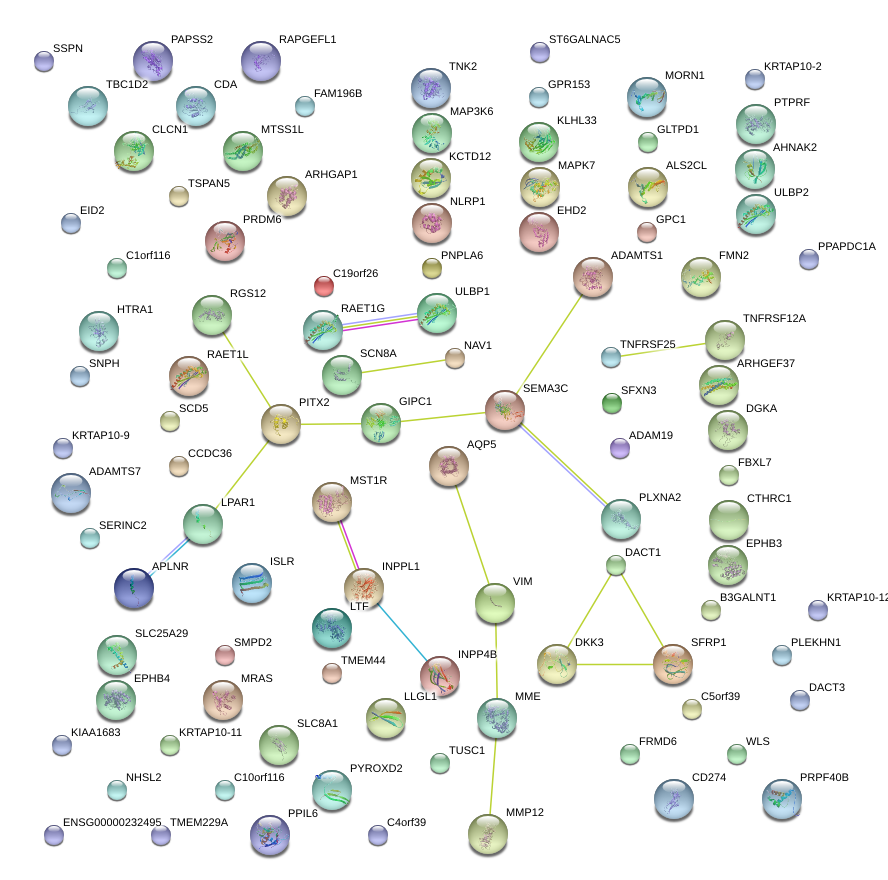
chr12_+_50271283
|
1.318
|
NM_014191
|
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit
|
|
chr10_+_17311962
|
1.314
|
|
VIM
|
vimentin
|
|
chr4_-_99798563
|
1.174
|
NM_005723
|
TSPAN5
|
tetraspanin 5
|
|
chr7_-_80386602
|
1.106
|
NM_006379
|
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C
|
|
chr3_+_185762233
|
1.046
|
NM_004443
|
EPHB3
|
EPH receptor B3
|
|
chr11_+_61491793
|
1.044
|
|
LOC399900
|
hypothetical LOC399900
|
|
chr7_-_80386258
|
1.024
|
|
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C
|
|
chr6_-_109868400
|
1.004
|
|
PPIL6
|
peptidylprolyl isomerase (cyclophilin)-like 6
|
|
chr2_+_241023703
|
0.997
|
NM_002081
|
GPC1
|
glypican 1
|
|
chr22_+_44828338
|
0.979
|
|
|
|
|
chr10_-_17311961
|
0.972
|
|
|
|
|
chr10_+_124210980
|
0.945
|
NM_002775
|
HTRA1
|
HtrA serine peptidase 1
|
|
chr1_+_199975674
|
0.933
|
|
NAV1
|
neuron navigator 1
|
|
chr5_-_169340321
|
0.917
|
NM_001129891
|
FAM196B
|
family with sequence similarity 196, member B
|
|
chr10_+_102782151
|
0.893
|
|
SFXN3
|
sideroflexin 3
|
|
chr1_+_199975563
|
0.891
|
NM_001167738
|
NAV1
|
neuron navigator 1
|
|
chr14_-_99842519
|
0.885
|
NM_001039355
|
SLC25A29
|
solute carrier family 25, member 29
|
|
chr5_+_148941228
|
0.866
|
NM_001001669
|
ARHGEF37
|
Rho guanine nucleotide exchange factor (GEF) 37
|
|
chr14_+_51025604
|
0.864
|
NM_001042481
|
FRMD6
|
FERM domain containing 6
|
|
chr1_-_6243621
|
0.863
|
NM_207370
|
GPR153
|
G protein-coupled receptor 153
|
|
chr15_-_76890626
|
0.858
|
NM_014272
|
ADAMTS7
|
ADAM metallopeptidase with thrombospondin type 1 motif, 7
|
|
chr9_-_21549696
|
0.853
|
|
LOC554202
|
hypothetical LOC554202
|
|
chr5_-_156935283
|
0.852
|
|
|
|
|
chr21_-_27139563
|
0.798
|
NM_006988
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1
|
|
chr12_+_54611212
|
0.786
|
NM_001345
|
DGKA
|
diacylglycerol kinase, alpha 80kDa
|
|
chr1_+_238321794
|
0.771
|
NM_020066
|
FMN2
|
formin 2
|
|
chr19_-_1188840
|
0.767
|
NM_152769
|
C19orf26
|
chromosome 19 open reading frame 26
|
|
chr12_+_26239755
|
0.751
|
NM_005086
|
SSPN
|
sarcospan (Kras oncogene-associated gene)
|
|
chr8_-_41286088
|
0.733
|
NM_003012
|
SFRP1
|
secreted frizzled-related protein 1
|
|
chr1_-_2312850
|
0.732
|
NM_024848
|
MORN1
|
MORN repeat containing 1
|
|
chr16_+_3010311
|
0.728
|
NM_016639
|
TNFRSF12A
|
tumor necrosis factor receptor superfamily, member 12A
|
|
chr4_-_143987053
|
0.723
|
NM_001101669
NM_003866
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa
|
|
chr12_+_26239535
|
0.715
|
NM_001135823
|
SSPN
|
sarcospan (Kras oncogene-associated gene)
|
|
chr3_-_49916304
|
0.714
|
NM_002447
|
MST1R
|
macrophage stimulating 1 receptor (c-met-related tyrosine kinase)
|
|
chr4_+_166097548
|
0.693
|
NM_153027
|
C4orf39
|
chromosome 4 open reading frame 39
|
|
chr1_-_206484258
|
0.689
|
NM_025179
|
PLXNA2
|
plexin A2
|
|
chr12_+_48641545
|
0.687
|
NM_001651
|
AQP5
|
aquaporin 5
|
|
chr5_+_122452739
|
0.682
|
NM_001136239
|
PRDM6
|
PR domain containing 6
|
|
chr1_-_27565412
|
0.681
|
|
MAP3K6
|
mitogen-activated protein kinase kinase kinase 6
|
|
chr6_-_150285838
|
0.675
|
NM_001001788
|
RAET1G
|
retinoic acid early transcript 1G
|
|
chr1_+_891739
|
0.673
|
NM_001160184
NM_032129
|
PLEKHN1
|
pleckstrin homology domain containing, family N member 1
|
|
chr21_+_44941514
|
0.673
|
NM_198699
|
KRTAP10-12
|
keratin associated protein 10-12
|
|
chr1_+_20788027
|
0.668
|
NM_001785
|
CDA
|
cytidine deaminase
|
|
chr5_-_156935317
|
0.662
|
NM_033274
|
ADAM19
|
ADAM metallopeptidase domain 19
|
|
chr15_+_72253940
|
0.661
|
NM_201526
|
ISLR
|
immunoglobulin superfamily containing leucine-rich repeat
|
|
chr11_+_71613472
|
0.640
|
NM_001567
|
INPPL1
|
inositol polyphosphate phosphatase-like 1
|
|
chr19_-_18246166
|
0.633
|
NM_001145304
NM_001145305
NM_025249
|
KIAA1683
|
KIAA1683
|
|
chr4_+_166097579
|
0.628
|
|
C4orf39
|
chromosome 4 open reading frame 39
|
|
chr7_-_100262971
|
0.627
|
NM_004444
|
EPHB4
|
EPH receptor B4
|
|
chr3_-_46710137
|
0.620
|
NM_001190707
NM_147129
|
ALS2CL
|
ALS2 C-terminal like
|
|
chr5_+_15553288
|
0.619
|
NM_012304
|
FBXL7
|
F-box and leucine-rich repeat protein 7
|
|
chr3_-_197103848
|
0.619
|
|
TNK2
|
tyrosine kinase, non-receptor, 2
|
|
chr10_-_100164877
|
0.617
|
NM_032709
|
PYROXD2
|
pyridine nucleotide-disulphide oxidoreductase domain 2
|
|
chr5_-_43075904
|
0.616
|
NM_001014279
|
C5orf39
|
chromosome 5 open reading frame 39
|
|
chr14_+_58174489
|
0.606
|
NM_001079520
NM_016651
|
DACT1
|
dapper, antagonist of beta-catenin, homolog 1 (Xenopus laevis)
|
|
chr1_+_43769065
|
0.605
|
NM_002840
NM_130440
|
PTPRF
|
protein tyrosine phosphatase, receptor type, F
|
|
chr9_-_100057809
|
0.597
|
NM_018421
|
TBC1D2
|
TBC1 domain family, member 2
|
|
chr1_+_1250152
|
0.595
|
|
|
|
|
chr9_-_112840801
|
0.588
|
|
LPAR1
|
lysophosphatidic acid receptor 1
|
|
chr5_+_140767953
|
0.577
|
NM_018926
NM_032100
|
PCDHGB6
|
protocadherin gamma subfamily B, 6
|
|
chr3_-_162305319
|
0.574
|
NM_033169
NM_001038628
NM_003781
NM_033167
NM_033168
|
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group)
|
|
chr9_-_25668230
|
0.573
|
|
TUSC1
|
tumor suppressor candidate 1
|
|
chr10_+_89409332
|
0.559
|
NM_001015880
NM_004670
|
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2
|
|
chr11_-_46678736
|
0.559
|
|
ARHGAP1
|
Rho GTPase activating protein 1
|
|
chr19_+_45807909
|
0.557
|
|
|
|
|
chr21_+_44871467
|
0.555
|
NM_198690
|
KRTAP10-9
|
keratin associated protein 10-9
|
|
chr19_+_7505037
|
0.550
|
NM_006702
NM_001166112
|
PNPLA6
|
patatin-like phospholipase domain containing 6
|
|
chr1_-_6448737
|
0.548
|
NM_001039664
NM_003790
NM_148965
NM_148966
NM_148967
NM_148970
|
TNFRSF25
|
tumor necrosis factor receptor superfamily, member 25
|
|
chr3_+_49211936
|
0.547
|
NM_001135197
|
CCDC36
|
coiled-coil domain containing 36
|
|
chr21_-_44795769
|
0.531
|
NM_198693
|
KRTAP10-2
|
keratin associated protein 10-2
|
|
chr9_-_100057313
|
0.530
|
|
TBC1D2
|
TBC1 domain family, member 2
|
|
chr21_-_27139143
|
0.524
|
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1
|
|
chrX_+_71047344
|
0.523
|
NM_001013627
|
NHSL2
|
NHS-like 2
|
|
chr14_-_104507862
|
0.513
|
|
AHNAK2
|
AHNAK nucleoprotein 2
|
|
chr3_-_46481346
|
0.508
|
NM_002343
|
LTF
|
lactotransferrin
|
|
chr7_-_123460722
|
0.504
|
NM_001136002
|
TMEM229A
|
transmembrane protein 229A
|
|
chr10_+_88718167
|
0.504
|
NM_006829
|
C10orf116
|
chromosome 10 open reading frame 116
|
|
chr17_-_5428491
|
0.504
|
NM_001033053
NM_014922
NM_033004
NM_033006
NM_033007
|
NLRP1
|
NLR family, pyrin domain containing 1
|
|
chr1_+_1250115
|
0.503
|
|
GLTPD1
|
glycolipid transfer protein domain containing 1
|
|
chr2_-_56266408
|
0.502
|
|
LOC100129434
|
hypothetical LOC100129434
|
|
chr11_-_46678691
|
0.501
|
NM_004308
|
ARHGAP1
|
Rho GTPase activating protein 1
|
|
chr17_+_18069593
|
0.501
|
NM_004140
|
LLGL1
|
lethal giant larvae homolog 1 (Drosophila)
|
|
chr10_+_122206455
|
0.500
|
NM_001030059
|
PPAPDC1A
|
phosphatidic acid phosphatase type 2 domain containing 1A
|
|
chr9_+_5440455
|
0.499
|
NM_014143
|
CD274
|
CD274 molecule
|
|
chr13_-_76358433
|
0.498
|
NM_138444
|
KCTD12
|
potassium channel tetramerisation domain containing 12
|
|
chr1_+_77105746
|
0.498
|
NM_030965
|
ST6GALNAC5
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5
|
|
chr7_+_142723340
|
0.493
|
NM_000083
|
CLCN1
|
chloride channel 1, skeletal muscle
|
|
chr5_+_15553539
|
0.489
|
|
FBXL7
|
F-box and leucine-rich repeat protein 7
|
|
chr12_+_54612055
|
0.481
|
NM_201444
NM_201445
NM_201554
|
DGKA
|
diacylglycerol kinase, alpha 80kDa
|
|
chr13_-_76358370
|
0.480
|
|
KCTD12
|
potassium channel tetramerisation domain containing 12
|
|
chr1_+_77105709
|
0.475
|
|
ST6GALNAC5
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5
|
|
chr11_-_11986704
|
0.472
|
NM_001018057
|
DKK3
|
dickkopf homolog 3 (Xenopus laevis)
|
|
chr4_+_3264509
|
0.466
|
|
RGS12
|
regulator of G-protein signaling 12
|
|
chr6_+_150326802
|
0.465
|
NM_025218
|
ULBP1
|
UL16 binding protein 1
|
|
chr9_-_25668855
|
0.457
|
NM_001004125
|
TUSC1
|
tumor suppressor candidate 1
|
|
chr3_-_195835197
|
0.452
|
|
TMEM44
|
transmembrane protein 44
|
|
chr8_+_104452961
|
0.447
|
NM_138455
|
CTHRC1
|
collagen triple helix repeat containing 1
|
|
chr20_+_1194959
|
0.447
|
NM_014723
|
SNPH
|
syntaphilin
|
|
chr6_+_109868736
|
0.446
|
|
SMPD2
|
sphingomyelin phosphodiesterase 2, neutral membrane (neutral sphingomyelinase)
|
|
chr14_-_19973640
|
0.446
|
NM_001109997
|
KLHL33
|
kelch-like 33 (Drosophila)
|
|
chr5_+_148766632
|
0.438
|
|
LOC728264
|
hypothetical LOC728264
|
|
chr5_+_15553768
|
0.434
|
|
FBXL7
|
F-box and leucine-rich repeat protein 7
|
|
chr1_-_68470585
|
0.432
|
NM_001002292
NM_001193334
NM_024911
|
WLS
|
wntless homolog (Drosophila)
|
|
chr5_+_15553736
|
0.431
|
|
FBXL7
|
F-box and leucine-rich repeat protein 7
|
|
chr2_+_241023908
|
0.429
|
|
GPC1
|
glypican 1
|
|
chr2_-_40532651
|
0.429
|
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1
|
|
chr19_+_52908412
|
0.429
|
NM_014601
|
EHD2
|
EH-domain containing 2
|
|
chr3_+_139550368
|
0.425
|
|
MRAS
|
muscle RAS oncogene homolog
|
|
chr16_-_69277355
|
0.418
|
NM_138383
|
MTSS1L
|
metastasis suppressor 1-like
|
|
chr19_-_44722599
|
0.412
|
NM_153232
|
EID2
|
EP300 interacting inhibitor of differentiation 2
|
|
chr10_+_17311788
|
0.410
|
|
VIM
|
vimentin
|
|
chr12_+_48303607
|
0.410
|
NM_012272
|
PRPF40B
|
PRP40 pre-mRNA processing factor 40 homolog B (S. cerevisiae)
|
|
chr10_+_124211353
|
0.408
|
|
HTRA1
|
HtrA serine peptidase 1
|
|
chr3_+_156280129
|
0.399
|
NM_000902
|
MME
|
membrane metallo-endopeptidase
|
|
chr1_-_205272619
|
0.399
|
NM_001083924
NM_023938
|
C1orf116
|
chromosome 1 open reading frame 116
|
|
chr11_-_56761250
|
0.396
|
NM_005161
|
APLNR
|
apelin receptor
|
|
chr17_+_19221606
|
0.392
|
NM_139032
NM_139033
|
MAPK7
|
mitogen-activated protein kinase 7
|
|
chr21_+_44890758
|
0.390
|
NM_198692
|
KRTAP10-11
|
keratin associated protein 10-11
|
|
chr4_-_83938920
|
0.387
|
NM_001037582
NM_024906
|
SCD5
|
stearoyl-CoA desaturase 5
|
|
chr19_-_14467939
|
0.385
|
NM_005716
NM_202467
NM_202468
NM_202469
NM_202470
NM_202494
|
GIPC1
|
GIPC PDZ domain containing family, member 1
|
|
chr1_+_31659246
|
0.384
|
NM_001199039
NM_018565
|
SERINC2
|
serine incorporator 2
|
|
chr17_+_35587767
|
0.384
|
NM_016339
|
RAPGEFL1
|
Rap guanine nucleotide exchange factor (GEF)-like 1
|
|
chr4_-_111777584
|
0.380
|
|
PITX2
|
paired-like homeodomain 2
|
|
chr16_-_674357
|
0.379
|
NM_001005920
|
JMJD8
|
jumonji domain containing 8
|
|
chr15_+_65145215
|
0.379
|
|
SMAD3
|
SMAD family member 3
|
|
chr12_+_106236281
|
0.377
|
NM_001018072
|
BTBD11
|
BTB (POZ) domain containing 11
|
|
chr12_+_6179736
|
0.373
|
NM_001769
|
CD9
|
CD9 molecule
|
|
chr22_+_38720881
|
0.373
|
NM_138435
|
FAM83F
|
family with sequence similarity 83, member F
|
|
chr3_-_195835298
|
0.371
|
NM_001011655
NM_001166305
NM_001166306
NM_138399
|
TMEM44
|
transmembrane protein 44
|
|
chr1_+_29011240
|
0.370
|
NM_000911
|
OPRD1
|
opioid receptor, delta 1
|
|
chr9_-_129656413
|
0.368
|
|
ENG
|
endoglin
|
|
chr5_+_34692239
|
0.364
|
|
RAI14
|
retinoic acid induced 14
|
|
chr11_-_118286822
|
0.363
|
NM_182557
|
BCL9L
|
B-cell CLL/lymphoma 9-like
|
|
chr19_-_4016467
|
0.363
|
|
ZBTB7A
|
zinc finger and BTB domain containing 7A
|
|
chr8_+_104452877
|
0.360
|
|
CTHRC1
|
collagen triple helix repeat containing 1
|
|
chr17_-_4831388
|
0.359
|
NM_001171167
NM_001171168
|
CAMTA2
|
calmodulin binding transcription activator 2
|
|
chrX_+_154264931
|
0.359
|
NM_001007523
NM_001007524
NM_012151
|
F8A2
F8A3
F8A1
|
coagulation factor VIII-associated (intronic transcript) 2
coagulation factor VIII-associated (intronic transcript) 3
coagulation factor VIII-associated (intronic transcript) 1
|
|
chr16_-_1086244
|
0.358
|
NM_207419
|
C1QTNF8
|
C1q and tumor necrosis factor related protein 8
|
|
chr1_+_199884072
|
0.356
|
NM_020443
|
NAV1
|
neuron navigator 1
|
|
chr5_+_94981735
|
0.355
|
NM_199243
|
GPR150
|
G protein-coupled receptor 150
|
|
chr11_-_67163600
|
0.353
|
NM_005995
|
TBX10
|
T-box 10
|
|
chr21_-_44836811
|
0.352
|
NM_198688
|
KRTAP10-6
|
keratin associated protein 10-6
|
|
chr3_-_188903038
|
0.351
|
NM_001004312
|
RTP2
|
receptor (chemosensory) transporter protein 2
|
|
chr1_+_199884152
|
0.351
|
|
NAV1
|
neuron navigator 1
|
|
chr17_+_17952744
|
0.349
|
NM_016239
|
MYO15A
|
myosin XVA
|
|
chr10_-_31360778
|
0.349
|
NM_001143766
NM_001143767
NM_001143768
NM_001143770
NM_001143771
NM_182755
|
ZNF438
|
zinc finger protein 438
|
|
chr4_+_148621518
|
0.349
|
NM_001166055
NM_001957
|
EDNRA
|
endothelin receptor type A
|
|
chr9_+_35595339
|
0.346
|
|
TESK1
|
testis-specific kinase 1
|
|
chr9_-_129656581
|
0.346
|
|
ENG
|
endoglin
|
|
chr1_+_156417302
|
0.345
|
|
CD1D
|
CD1d molecule
|
|
chr2_+_17923404
|
0.344
|
NM_002252
|
KCNS3
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3
|
|
chr17_-_40750109
|
0.344
|
NM_003954
|
MAP3K14
|
mitogen-activated protein kinase kinase kinase 14
|
|
chr15_+_65145248
|
0.342
|
NM_005902
|
SMAD3
|
SMAD family member 3
|
|
chr1_+_115198977
|
0.342
|
NM_003176
|
SYCP1
|
synaptonemal complex protein 1
|
|
chr3_-_50371934
|
0.341
|
NM_007024
|
TMEM115
|
transmembrane protein 115
|
|
chr19_-_14467890
|
0.341
|
|
GIPC1
|
GIPC PDZ domain containing family, member 1
|
|
chr11_-_68537243
|
0.341
|
NM_001098515
NM_145015
|
MRGPRF
|
MAS-related GPR, member F
|
|
chr21_+_32706615
|
0.339
|
NM_058187
|
C21orf63
|
chromosome 21 open reading frame 63
|
|
chr4_-_25473528
|
0.338
|
NM_015187
|
SEL1L3
|
sel-1 suppressor of lin-12-like 3 (C. elegans)
|
|
chr12_-_46439127
|
0.336
|
NM_001098531
|
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3
|
|
chr8_-_142307854
|
0.335
|
NM_001080431
|
SLC45A4
|
solute carrier family 45, member 4
|
|
chr5_-_33928049
|
0.335
|
|
ADAMTS12
|
ADAM metallopeptidase with thrombospondin type 1 motif, 12
|
|
chr17_+_21220260
|
0.333
|
NM_021012
|
KCNJ12
|
potassium inwardly-rectifying channel, subfamily J, member 12
|
|
chr17_-_76064842
|
0.331
|
NM_002522
|
NPTX1
|
neuronal pentraxin I
|
|
chr21_+_33364316
|
0.329
|
NM_138983
|
OLIG1
|
oligodendrocyte transcription factor 1
|
|
chr5_+_34692352
|
0.329
|
NM_001145520
|
RAI14
|
retinoic acid induced 14
|
|
chr1_-_158182006
|
0.328
|
NM_001135050
NM_020789
|
IGSF9
|
immunoglobulin superfamily, member 9
|
|
chr2_-_222871577
|
0.328
|
NM_000438
NM_001127366
NM_013942
NM_181457
NM_181458
NM_181459
NM_181460
NM_181461
|
PAX3
|
paired box 3
|
|
chr1_-_150233337
|
0.325
|
NM_002966
|
S100A10
|
S100 calcium binding protein A10
|
|
chr5_+_34692124
|
0.325
|
NM_001145522
NM_015577
|
RAI14
|
retinoic acid induced 14
|
|
chr3_-_50371896
|
0.325
|
|
TMEM115
|
transmembrane protein 115
|
|
chr4_-_140443061
|
0.324
|
NM_001184987
NM_001184988
NM_001184989
NM_001184990
|
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa
|
|
chr17_+_6495694
|
0.320
|
NM_001105520
|
C17orf100
|
chromosome 17 open reading frame 100
|
|
chr9_+_35595275
|
0.317
|
NM_006285
|
TESK1
|
testis-specific kinase 1
|
|
chr9_-_129707392
|
0.317
|
|
LOC100131355
ST6GALNAC6
|
hypothetical protein LOC100131355
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6
|
|
chr10_+_11824361
|
0.316
|
NM_024693
|
ECHDC3
|
enoyl CoA hydratase domain containing 3
|
|
chr8_+_143778529
|
0.316
|
NM_001160354
NM_001160355
NM_017527
|
LY6K
|
lymphocyte antigen 6 complex, locus K
|
|
chr16_+_2016807
|
0.314
|
NM_001130012
NM_004785
|
SLC9A3R2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2
|
|
chr9_-_129656778
|
0.313
|
|
ENG
|
endoglin
|
|
chr5_-_157011949
|
0.311
|
NM_007017
NM_178424
|
SOX30
|
SRY (sex determining region Y)-box 30
|
|
chr17_+_19222651
|
0.309
|
|
MAPK7
|
mitogen-activated protein kinase 7
|
|
chrX_+_106955737
|
0.308
|
NM_012216
NM_052817
|
MID2
|
midline 2
|
|
chr3_-_50371960
|
0.305
|
|
TMEM115
|
transmembrane protein 115
|
|
chr3_+_129117007
|
0.301
|
|
KBTBD12
|
kelch repeat and BTB (POZ) domain containing 12
|
|
chr2_-_232103352
|
0.301
|
NM_006056
|
NMUR1
|
neuromedin U receptor 1
|
|
chr17_+_70495318
|
0.299
|
NM_014603
|
CDR2L
|
cerebellar degeneration-related protein 2-like
|
|
chr22_-_16020153
|
0.298
|
|
CECR5
|
cat eye syndrome chromosome region, candidate 5
|
|
chr11_-_64284762
|
0.297
|
NM_001164716
NM_005609
|
PYGM
|
phosphorylase, glycogen, muscle
|
|
chr9_+_129951552
|
0.297
|
NM_005564
|
LCN2
|
lipocalin 2
|
|
chr11_+_696607
|
0.297
|
|
EPS8L2
|
EPS8-like 2
|
|
chr10_+_73393998
|
0.295
|
NM_004273
|
CHST3
|
carbohydrate (chondroitin 6) sulfotransferase 3
|
|
chr1_+_208568811
|
0.295
|
NM_001170587
NM_001170588
NM_018194
|
HHAT
|
hedgehog acyltransferase
|
|
chr19_-_59385226
|
0.294
|
|
MBOAT7
|
membrane bound O-acyltransferase domain containing 7
|
|
chr15_-_70455153
|
0.293
|
|
HEXA
|
hexosaminidase A (alpha polypeptide)
|
|
chr3_+_158026769
|
0.292
|
NM_001004316
NM_001193283
|
LEKR1
|
leucine, glutamate and lysine rich 1
|
|
chr1_+_31658549
|
0.291
|
NM_178865
|
SERINC2
|
serine incorporator 2
|
|
chr22_-_16020168
|
0.290
|
NM_033070
|
CECR5
|
cat eye syndrome chromosome region, candidate 5
|
|
chr6_+_86216020
|
0.290
|
NM_002526
|
NT5E
|
5'-nucleotidase, ecto (CD73)
|
|
chr5_-_157011665
|
0.289
|
|
|
|
|
chr1_-_150232900
|
0.289
|
|
S100A10
|
S100 calcium binding protein A10
|