|
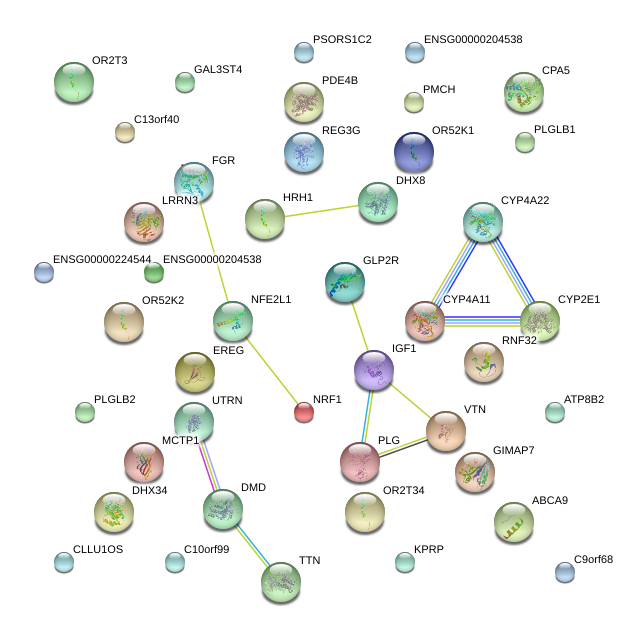
chr12_-_91346054
|
0.115
|
NM_001025232
|
CLLU1OS
|
chronic lymphocytic leukemia up-regulated 1 opposite strand
|
|
chr8_+_39911630
|
0.095
|
NM_194294
|
IDO2
|
indoleamine 2,3-dioxygenase 2
|
|
chr1_+_246703249
|
0.078
|
NM_001005495
|
OR2T3
|
olfactory receptor, family 2, subfamily T, member 3
|
|
chr6_-_31215089
|
0.062
|
NM_014069
|
PSORS1C2
|
psoriasis susceptibility 1 candidate 2
|
|
chr1_-_27825178
|
0.061
|
NM_001042747
|
FGR
|
Gardner-Rasheed feline sarcoma viral (v-fgr) oncogene homolog
|
|
chr3_-_44464722
|
0.053
|
|
|
|
|
chr7_+_129771865
|
0.048
|
NM_001127441
NM_001127442
NM_080385
|
CPA5
|
carboxypeptidase A5
|
|
chr2_-_179380301
|
0.044
|
NM_003319
NM_133378
NM_133379
NM_133432
NM_133437
|
TTN
|
titin
|
|
chr1_+_556968
|
0.041
|
|
|
|
|
chr10_+_85923473
|
0.040
|
NM_207373
|
C10orf99
|
chromosome 10 open reading frame 99
|
|
chr4_+_75449720
|
0.036
|
NM_001432
|
EREG
|
epiregulin
|
|
chr2_+_79106333
|
0.035
|
NM_001008387
NM_198448
|
REG3G
|
regenerating islet-derived 3 gamma
|
|
chr11_+_4466684
|
0.026
|
NM_001005171
|
OR52K1
|
olfactory receptor, family 52, subfamily K, member 1
|
|
chr7_+_156128431
|
0.024
|
NM_001184997
|
RNF32
|
ring finger protein 32
|
|
chr12_-_101115677
|
0.024
|
NM_002674
|
PMCH
|
pro-melanin-concentrating hormone
|
|
chr7_-_99604171
|
0.023
|
NM_024637
|
GAL3ST4
|
galactose-3-O-sulfotransferase 4
|
|
chr1_-_246804680
|
0.022
|
NM_001001821
|
OR2T34
|
olfactory receptor, family 2, subfamily T, member 34
|
|
chr7_+_110518297
|
0.021
|
NM_001099658
NM_001099660
NM_018334
|
LRRN3
|
leucine rich repeat neuronal 3
|
|
chr2_-_87102453
|
0.020
|
NM_002665
NM_001032392
|
PLGLB2
PLGLB1
|
plasminogen-like B2
plasminogen-like B1
|
|
chr12_-_101396459
|
0.019
|
NM_001111284
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr17_-_64568641
|
0.019
|
NM_080283
|
ABCA9
|
ATP-binding cassette, sub-family A (ABC1), member 9
|
|
chr17_+_9686572
|
0.018
|
|
GLP2R
|
glucagon-like peptide 2 receptor
|
|
chr7_+_149842850
|
0.014
|
NM_153236
|
GIMAP7
|
GTPase, IMAP family member 7
|
|
chr1_+_150997129
|
0.014
|
NM_001025231
|
KPRP
|
keratinocyte proline-rich protein
|
|
chr3_+_11153778
|
0.011
|
NM_001098213
|
HRH1
|
histamine receptor H1
|
|
chrX_-_33139349
|
0.008
|
NM_004006
|
DMD
|
dystrophin
|
|
chr1_+_152566899
|
0.007
|
NM_020452
|
ATP8B2
|
ATPase, class I, type 8B, member 2
|
|
chr1_+_66570683
|
0.006
|
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific
|
|
chr7_+_129057152
|
0.006
|
NM_001040110
|
NRF1
|
nuclear respiratory factor 1
|
|
chr17_-_23721485
|
0.005
|
NM_000638
|
VTN
|
vitronectin
|
|
chr1_-_47179729
|
0.004
|
NM_000778
|
CYP4A11
CYP4A22
|
cytochrome P450, family 4, subfamily A, polypeptide 11
cytochrome P450, family 4, subfamily A, polypeptide 22
|
|
chr9_-_4656665
|
0.004
|
NM_001039395
|
C9orf68
|
chromosome 9 open reading frame 68
|
|
chr13_-_102209422
|
0.003
|
NM_001146197
|
C13orf40
|
chromosome 13 open reading frame 40
|
|
chr10_+_135190855
|
0.003
|
NM_000773
|
CYP2E1
|
cytochrome P450, family 2, subfamily E, polypeptide 1
|
|
chr5_-_94250357
|
0.002
|
|
MCTP1
|
multiple C2 domains, transmembrane 1
|