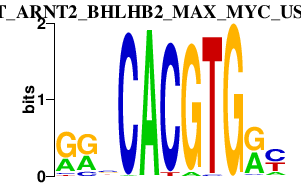
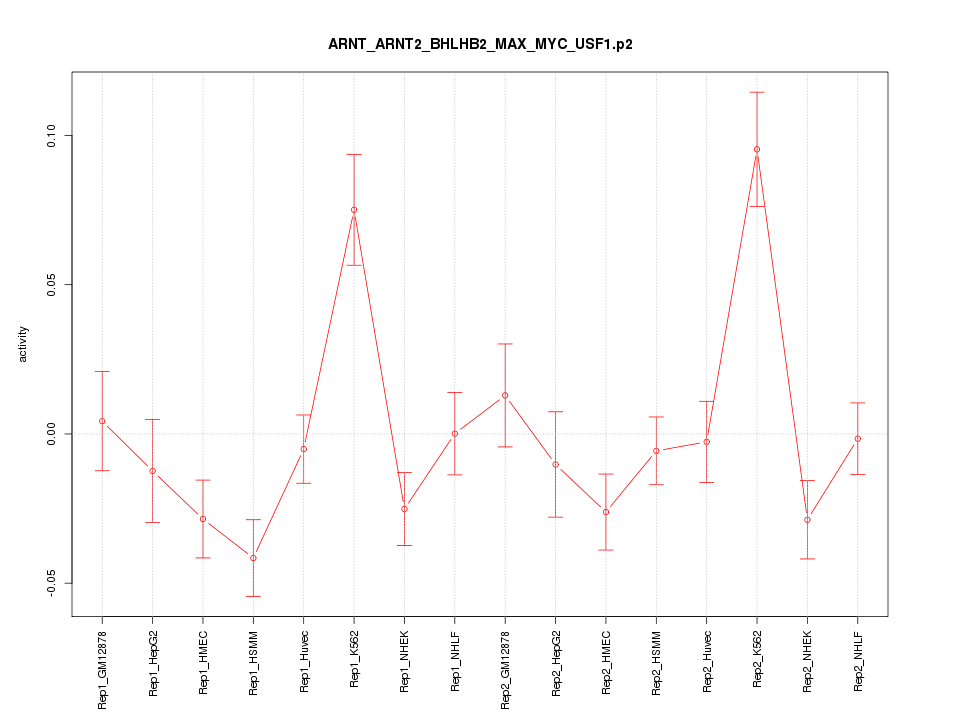
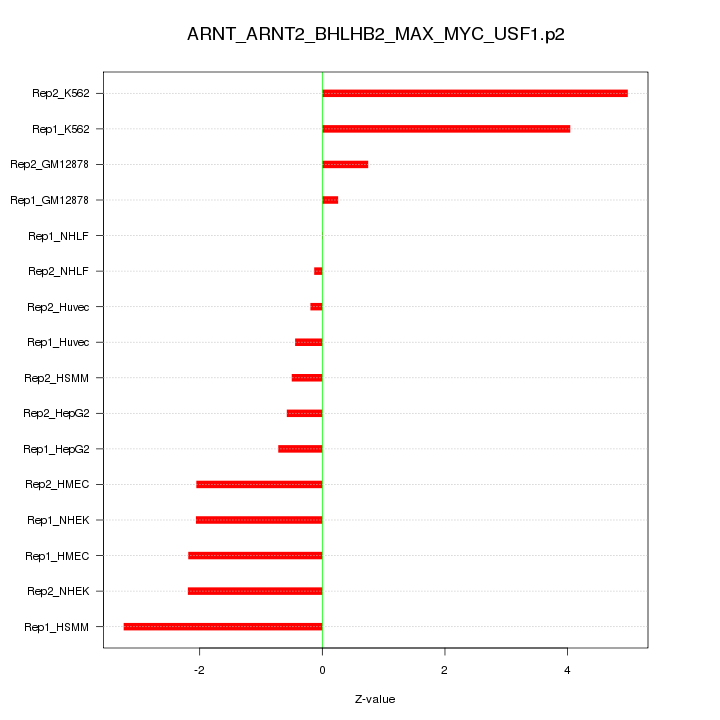
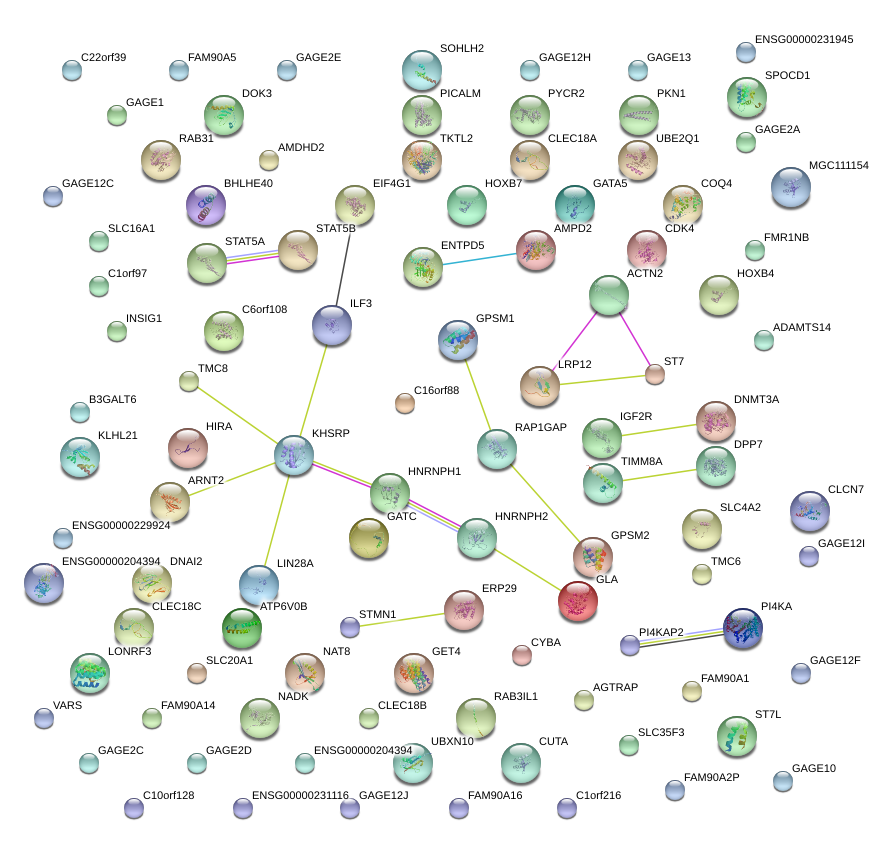
|
chr16_+_2510323
|
6.142
|
NM_001145815
NM_015944
|
AMDHD2
|
amidohydrolase domain containing 2
|
|
chr17_-_53764867
|
6.080
|
|
|
|
|
chr1_+_20385164
|
5.245
|
NM_152376
|
UBXN10
|
UBX domain protein 10
|
|
chr12_-_56432292
|
4.023
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr19_+_10625896
|
3.836
|
NM_001137673
NM_004516
NM_012218
NM_017620
NM_153464
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa
|
|
chr19_+_10625987
|
3.835
|
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa
|
|
chr11_-_61441536
|
3.805
|
NM_013401
|
RAB3IL1
|
RAB3A interacting protein (rabin3)-like 1
|
|
chr7_+_150390566
|
3.758
|
|
SLC4A2
|
solute carrier family 4, anion exchanger, member 2 (erythrocyte membrane protein band 3-like 1)
|
|
chr12_-_56432377
|
3.757
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr12_-_56432375
|
3.691
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr12_-_56432385
|
3.580
|
NM_000075
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr16_-_1465021
|
3.487
|
|
CLCN7
|
chloride channel 7
|
|
chrX_+_100549869
|
3.486
|
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H')
|
|
chrX_+_100549861
|
3.305
|
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H')
|
|
chr19_+_10626106
|
3.260
|
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa
|
|
chrX_+_100549839
|
3.256
|
NM_001032393
NM_019597
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H')
|
|
chr9_-_139128983
|
2.978
|
|
DPP7
|
dipeptidyl-peptidase 7
|
|
chrX_+_100549885
|
2.912
|
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H')
|
|
chr9_-_139128777
|
2.897
|
|
DPP7
|
dipeptidyl-peptidase 7
|
|
chrX_-_100549561
|
2.868
|
NM_000169
|
GLA
|
galactosidase, alpha
|
|
chr6_+_160310118
|
2.809
|
NM_000876
|
IGF2R
|
insulin-like growth factor 2 receptor
|
|
chr16_-_1465007
|
2.757
|
|
CLCN7
|
chloride channel 7
|
|
chr6_-_31871394
|
2.742
|
|
VARS
|
valyl-tRNA synthetase
|
|
chr10_+_72102563
|
2.643
|
NM_080722
NM_139155
|
ADAMTS14
|
ADAM metallopeptidase with thrombospondin type 1 motif, 14
|
|
chr11_-_71492019
|
2.630
|
NM_017907
|
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1
|
|
chr1_-_224178600
|
2.599
|
|
PYCR2
|
pyrroline-5-carboxylate reductase family, member 2
|
|
chr19_-_10625449
|
2.491
|
|
LOC147727
|
hypothetical LOC147727
|
|
chr1_-_224178606
|
2.459
|
|
PYCR2
|
pyrroline-5-carboxylate reductase family, member 2
|
|
chr11_-_85457469
|
2.413
|
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr16_-_87244896
|
2.374
|
NM_000101
|
CYBA
|
cytochrome b-245, alpha polypeptide
|
|
chr2_-_25328452
|
2.354
|
NM_153759
|
DNMT3A
|
DNA (cytosine-5-)-methyltransferase 3 alpha
|
|
chr17_-_44010543
|
2.347
|
NM_024015
|
HOXB4
|
homeobox B4
|
|
chr16_-_1465041
|
2.347
|
NM_001114331
NM_001287
|
CLCN7
|
chloride channel 7
|
|
chr6_-_33494042
|
2.316
|
NM_001014433
NM_001014837
NM_001014838
NM_001014840
NM_015921
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli)
|
|
chr11_-_85457736
|
2.315
|
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr6_-_33493898
|
2.283
|
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli)
|
|
chr6_-_43305156
|
2.270
|
NM_006443
NM_199184
|
C6orf108
|
chromosome 6 open reading frame 108
|
|
chrX_+_146870540
|
2.256
|
NM_152578
|
FMR1NB
|
fragile X mental retardation 1 neighbor
|
|
chr6_-_33493792
|
2.253
|
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli)
|
|
chr1_-_1700088
|
2.225
|
NM_001198993
|
NADK
|
NAD kinase
|
|
chr1_-_152797820
|
2.223
|
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|
|
chr11_-_71491983
|
2.209
|
|
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1
|
|
chr16_+_68542310
|
2.198
|
NM_182619
|
CLEC18C
CLEC18A
CLEC18B
|
C-type lectin domain family 18, member C
C-type lectin domain family 18, member A
C-type lectin domain family 18, member B
|
|
chr16_+_68542126
|
2.175
|
NM_001136214
|
CLEC18A
|
C-type lectin domain family 18, member A
|
|
chr17_+_73638430
|
2.174
|
NM_152468
|
TMC8
|
transmembrane channel-like 8
|
|
chr11_-_85457753
|
2.163
|
NM_001008660
NM_007166
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr19_+_14405100
|
2.154
|
NM_002741
|
PKN1
|
protein kinase N1
|
|
chr1_+_1157485
|
2.150
|
NM_080605
|
B3GALT6
|
UDP-Gal:betaGal beta 1,3-galactosyltransferase polypeptide 6
|
|
chrX_-_100549444
|
2.150
|
|
GLA
|
galactosidase, alpha
|
|
chr20_-_60484420
|
2.140
|
NM_080473
|
GATA5
|
GATA binding protein 5
|
|
chr1_-_224178553
|
2.114
|
NM_013328
|
PYCR2
|
pyrroline-5-carboxylate reductase family, member 2
|
|
chr1_+_109963950
|
2.103
|
NM_139156
|
AMPD2
|
adenosine monophosphate deaminase 2
|
|
chr19_+_14405216
|
2.079
|
|
PKN1
|
protein kinase N1
|
|
chr11_-_71491942
|
2.072
|
|
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1
|
|
chrX_-_100490329
|
2.046
|
NM_001145951
|
TIMM8A
|
translocase of inner mitochondrial membrane 8 homolog A (yeast)
|
|
chr8_+_142471290
|
2.036
|
|
|
|
|
chr4_-_164614335
|
2.026
|
NM_032136
|
TKTL2
|
transketolase-like 2
|
|
chr12_+_110935618
|
2.017
|
|
ERP29
|
endoplasmic reticulum protein 29
|
|
chr17_-_73636362
|
1.986
|
|
TMC6
|
transmembrane channel-like 6
|
|
chr22_-_19543099
|
1.976
|
|
PI4KA
|
phosphatidylinositol 4-kinase, catalytic, alpha
|
|
chr1_-_35957030
|
1.953
|
|
C1orf216
|
chromosome 1 open reading frame 216
|
|
chr1_-_224178486
|
1.947
|
|
PYCR2
|
pyrroline-5-carboxylate reductase family, member 2
|
|
chr1_+_44213200
|
1.929
|
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chr12_-_8271460
|
1.920
|
NM_018088
|
FAM90A1
|
family with sequence similarity 90, member A1
|
|
chr1_+_109963982
|
1.918
|
|
AMPD2
|
adenosine monophosphate deaminase 2
|
|
chr18_+_9698299
|
1.909
|
|
RAB31
|
RAB31, member RAS oncogene family
|
|
chr1_-_152797760
|
1.903
|
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|
|
chr5_-_176869943
|
1.902
|
NM_001144875
NM_001144876
|
DOK3
|
docking protein 3
|
|
chr10_-_50066386
|
1.875
|
NM_001010863
|
C10orf128
|
chromosome 10 open reading frame 128
|
|
chr9_-_139128979
|
1.855
|
|
DPP7
|
dipeptidyl-peptidase 7
|
|
chr16_-_1464927
|
1.851
|
|
CLCN7
|
chloride channel 7
|
|
chrX_+_49241061
|
1.851
|
NM_001127212
|
GAGE8
GAGE1
GAGE2C
GAGE2A
|
G antigen 8
G antigen 1
G antigen 2C
G antigen 2A
|
|
chr1_-_152797767
|
1.844
|
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|
|
chr13_-_35686665
|
1.829
|
NM_017826
|
SOHLH2
|
spermatogenesis and oogenesis specific basic helix-loop-helix 2
|
|
chrX_+_117992603
|
1.825
|
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3
|
|
chrX_-_100490612
|
1.818
|
NM_004085
|
TIMM8A
|
translocase of inner mitochondrial membrane 8 homolog A (yeast)
|
|
chr1_-_26105954
|
1.808
|
NM_203399
|
STMN1
|
stathmin 1
|
|
chr1_+_44213229
|
1.807
|
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chr1_-_152797707
|
1.805
|
NM_017582
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|
|
chr17_+_37694007
|
1.804
|
|
STAT5A
|
signal transducer and activator of transcription 5A
|
|
chr1_+_44213182
|
1.796
|
NM_001039457
NM_004047
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chr7_+_882704
|
1.792
|
NM_015949
|
GET4
|
golgi to ER traffic protein 4 homolog (S. cerevisiae)
|
|
chr9_-_139129015
|
1.773
|
NM_013379
|
DPP7
|
dipeptidyl-peptidase 7
|
|
chr3_+_185515108
|
1.760
|
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1
|
|
chr7_+_116380590
|
1.760
|
NM_018412
NM_021908
|
ST7
|
suppression of tumorigenicity 7
|
|
chr7_+_154720416
|
1.750
|
NM_005542
NM_198336
NM_198337
|
INSIG1
|
insulin induced gene 1
|
|
chr3_+_4996141
|
1.732
|
|
BHLHE40
|
basic helix-loop-helix family, member e40
|
|
chr2_+_113119985
|
1.715
|
NM_005415
|
SLC20A1
|
solute carrier family 20 (phosphate transporter), member 1
|
|
chr9_-_139128992
|
1.714
|
|
DPP7
|
dipeptidyl-peptidase 7
|
|
chr3_+_4996305
|
1.704
|
|
BHLHE40
|
basic helix-loop-helix family, member e40
|
|
chr6_-_31871452
|
1.689
|
|
VARS
|
valyl-tRNA synthetase
|
|
chr1_-_113300138
|
1.682
|
NM_001166496
|
SLC16A1
|
solute carrier family 16, member 1 (monocarboxylic acid transporter 1)
|
|
chr22_-_17799177
|
1.674
|
|
HIRA
|
HIR histone cell cycle regulation defective homolog A (S. cerevisiae)
|
|
chr1_-_224178475
|
1.674
|
|
PYCR2
|
pyrroline-5-carboxylate reductase family, member 2
|
|
chr18_+_9698269
|
1.669
|
|
RAB31
|
RAB31, member RAS oncogene family
|
|
chr1_+_44213210
|
1.666
|
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chr1_+_109221271
|
1.663
|
|
GPSM2
|
G-protein signaling modulator 2
|
|
chr19_-_6375804
|
1.657
|
|
KHSRP
|
KH-type splicing regulatory protein
|
|
chr1_-_32054164
|
1.651
|
NM_144569
|
SPOCD1
|
SPOC domain containing 1
|
|
chr17_+_69781980
|
1.651
|
NM_001172810
NM_023036
|
DNAI2
|
dynein, axonemal, intermediate chain 2
|
|
chr9_-_139128990
|
1.648
|
|
DPP7
|
dipeptidyl-peptidase 7
|
|
chr1_-_21868380
|
1.641
|
NM_001145657
NM_002885
|
RAP1GAP
|
RAP1 GTPase activating protein
|
|
chr3_+_4996096
|
1.640
|
NM_003670
|
BHLHE40
|
basic helix-loop-helix family, member e40
|
|
chr22_-_19543012
|
1.635
|
|
PI4KA
|
phosphatidylinositol 4-kinase, catalytic, alpha
|
|
chr1_-_35956979
|
1.635
|
|
C1orf216
|
chromosome 1 open reading frame 216
|
|
chr1_-_26105477
|
1.630
|
NM_001145454
NM_005563
|
STMN1
|
stathmin 1
|
|
chr7_+_116380790
|
1.618
|
|
ST7
|
suppression of tumorigenicity 7
|
|
chr1_+_11718798
|
1.608
|
|
AGTRAP
|
angiotensin II receptor-associated protein
|
|
chr9_+_130124566
|
1.600
|
NM_016035
|
COQ4
|
coenzyme Q4 homolog (S. cerevisiae)
|
|
chr22_-_17799215
|
1.593
|
NM_003325
|
HIRA
|
HIR histone cell cycle regulation defective homolog A (S. cerevisiae)
|
|
chr17_-_44042924
|
1.591
|
|
HOXB7
|
homeobox B7
|
|
chr1_-_113300351
|
1.586
|
NM_003051
|
SLC16A1
|
solute carrier family 16, member 1 (monocarboxylic acid transporter 1)
|
|
chr9_-_139128988
|
1.581
|
|
DPP7
|
dipeptidyl-peptidase 7
|
|
chrX_+_117992623
|
1.579
|
NM_001031855
NM_024778
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3
|
|
chr1_+_26609819
|
1.565
|
NM_024674
|
LIN28A
|
lin-28 homolog A (C. elegans)
|
|
chr3_+_185515013
|
1.559
|
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1
|
|
chr1_+_234916376
|
1.558
|
NM_001103
|
ACTN2
|
actinin, alpha 2
|
|
chr1_-_6585268
|
1.547
|
|
KLHL21
|
kelch-like 21 (Drosophila)
|
|
chr2_-_80384454
|
1.538
|
|
LRRTM1
|
leucine rich repeat transmembrane neuronal 1
|
|
chr1_-_224178412
|
1.535
|
|
PYCR2
|
pyrroline-5-carboxylate reductase family, member 2
|
|
chr1_+_44213260
|
1.534
|
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chr22_+_18484810
|
1.526
|
NM_002882
|
RANBP1
|
RAN binding protein 1
|
|
chr17_-_59932698
|
1.519
|
|
DDX5
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 5
|
|
chr6_-_44333002
|
1.516
|
NM_178148
|
SLC35B2
|
solute carrier family 35, member B2
|
|
chr1_+_44870223
|
1.514
|
|
RNF220
|
ring finger protein 220
|
|
chrX_+_49075036
|
1.511
|
NM_012196
NM_001127200
NM_001098412
|
GAGE8
GAGE2C
GAGE2E
GAGE13
GAGE2A
|
G antigen 8
G antigen 2C
G antigen 2E
G antigen 13
G antigen 2A
|
|
chr12_-_107551783
|
1.506
|
NM_003006
|
SELPLG
|
selectin P ligand
|
|
chr1_+_152459918
|
1.499
|
NM_014847
|
UBAP2L
|
ubiquitin associated protein 2-like
|
|
chr1_+_11718715
|
1.495
|
NM_001040194
NM_001040195
NM_001040196
NM_001040197
NM_020350
|
AGTRAP
|
angiotensin II receptor-associated protein
|
|
chr1_-_205186330
|
1.494
|
NM_002644
|
PIGR
|
polymeric immunoglobulin receptor
|
|
chr1_-_11788666
|
1.490
|
NM_005957
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H)
|
|
chr3_+_195688974
|
1.486
|
|
FLJ34208
|
hypothetical LOC401106
|
|
chr1_-_40335496
|
1.483
|
|
PPT1
|
palmitoyl-protein thioesterase 1
|
|
chr22_+_18485057
|
1.481
|
|
RANBP1
|
RAN binding protein 1
|
|
chr16_+_2807164
|
1.478
|
NM_006799
NM_144956
NM_144957
|
PRSS21
|
protease, serine, 21 (testisin)
|
|
chr3_-_4483952
|
1.465
|
|
SUMF1
|
sulfatase modifying factor 1
|
|
chr12_+_110935612
|
1.459
|
|
|
|
|
chr1_-_11788550
|
1.459
|
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H)
|
|
chr1_-_40335451
|
1.454
|
|
PPT1
|
palmitoyl-protein thioesterase 1
|
|
chr22_+_18485076
|
1.446
|
|
RANBP1
|
RAN binding protein 1
|
|
chr19_+_10392128
|
1.437
|
NM_001111307
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chrX_+_49122651
|
1.433
|
NM_001098411
|
GAGE2B
|
G antigen 2B
|
|
chr16_+_2807228
|
1.427
|
|
PRSS21
|
protease, serine, 21 (testisin)
|
|
chr1_+_11718764
|
1.415
|
|
AGTRAP
|
angiotensin II receptor-associated protein
|
|
chr12_-_107551694
|
1.412
|
|
SELPLG
|
selectin P ligand
|
|
chr1_+_111544469
|
1.408
|
|
|
|
|
chr1_-_40335510
|
1.397
|
|
PPT1
|
palmitoyl-protein thioesterase 1
|
|
chr20_-_32877029
|
1.393
|
NM_014071
|
NCOA6
|
nuclear receptor coactivator 6
|
|
chr1_-_40335504
|
1.392
|
|
PPT1
|
palmitoyl-protein thioesterase 1
|
|
chr6_-_35764588
|
1.391
|
|
FKBP5
|
FK506 binding protein 5
|
|
chr19_-_6375782
|
1.383
|
|
KHSRP
|
KH-type splicing regulatory protein
|
|
chr12_+_110935644
|
1.378
|
|
ERP29
|
endoplasmic reticulum protein 29
|
|
chr22_+_18485068
|
1.376
|
|
RANBP1
|
RAN binding protein 1
|
|
chr22_-_22511173
|
1.373
|
NM_001002862
NM_001135751
NM_198440
|
DERL3
|
Der1-like domain family, member 3
|
|
chr16_+_88169612
|
1.364
|
NM_014427
NM_153636
|
CPNE7
|
copine VII
|
|
chr6_-_28999388
|
1.361
|
|
TRIM27
|
tripartite motif containing 27
|
|
chr12_-_107551744
|
1.356
|
|
SELPLG
|
selectin P ligand
|
|
chr17_-_44043278
|
1.354
|
|
HOXB7
|
homeobox B7
|
|
chrX_+_49094059
|
1.348
|
NM_012196
NM_001472
NM_001127200
NM_001098407
NM_001127212
|
GAGE8
GAGE2C
GAGE2E
GAGE2D
GAGE2A
|
G antigen 8
G antigen 2C
G antigen 2E
G antigen 2D
G antigen 2A
|
|
chr17_+_37941476
|
1.336
|
NM_000263
|
NAGLU
|
N-acetylglucosaminidase, alpha
|
|
chr6_-_35764655
|
1.335
|
NM_001145776
NM_001145777
NM_004117
|
FKBP5
|
FK506 binding protein 5
|
|
chr21_+_37367432
|
1.334
|
NM_001001894
|
TTC3
|
tetratricopeptide repeat domain 3
|
|
chr1_-_28432095
|
1.332
|
NM_014280
|
DNAJC8
|
DnaJ (Hsp40) homolog, subfamily C, member 8
|
|
chr12_+_88626770
|
1.332
|
|
LOC338758
|
hypothetical LOC338758
|
|
chr17_-_37586716
|
1.331
|
NM_012285
|
KCNH4
|
potassium voltage-gated channel, subfamily H (eag-related), member 4
|
|
chr5_+_43156741
|
1.331
|
|
ZNF131
|
zinc finger protein 131
|
|
chr7_+_106472243
|
1.324
|
NM_002736
|
PRKAR2B
|
protein kinase, cAMP-dependent, regulatory, type II, beta
|
|
chr17_+_37941790
|
1.317
|
|
NAGLU
|
N-acetylglucosaminidase, alpha
|
|
chr1_-_28432060
|
1.315
|
|
DNAJC8
|
DnaJ (Hsp40) homolog, subfamily C, member 8
|
|
chr16_-_87570716
|
1.314
|
NM_005187
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chr2_+_143603439
|
1.312
|
|
ARHGAP15
|
Rho GTPase activating protein 15
|
|
chr22_-_17798998
|
1.306
|
|
HIRA
|
HIR histone cell cycle regulation defective homolog A (S. cerevisiae)
|
|
chr22_+_18485145
|
1.301
|
|
RANBP1
|
RAN binding protein 1
|
|
chr3_-_4483928
|
1.295
|
|
SUMF1
|
sulfatase modifying factor 1
|
|
chr15_-_48766252
|
1.292
|
|
TRPM7
|
transient receptor potential cation channel, subfamily M, member 7
|
|
chr1_+_117098579
|
1.289
|
NM_001767
|
CD2
|
CD2 molecule
|
|
chr3_-_4483955
|
1.284
|
NM_001164674
NM_001164675
NM_182760
|
SUMF1
|
sulfatase modifying factor 1
|
|
chr1_-_11042668
|
1.278
|
NM_003132
|
SRM
|
spermidine synthase
|
|
chr1_-_227828416
|
1.278
|
NM_001025247
NM_014409
|
TAF5L
|
TAF5-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor, 65kDa
|
|
chr17_-_73636386
|
1.277
|
NM_001127198
|
TMC6
|
transmembrane channel-like 6
|
|
chr7_-_99536929
|
1.276
|
|
MCM7
|
minichromosome maintenance complex component 7
|
|
chr12_+_110935534
|
1.270
|
NM_001034025
NM_006817
|
ERP29
|
endoplasmic reticulum protein 29
|
|
chr19_-_6375820
|
1.265
|
NM_003685
|
KHSRP
|
KH-type splicing regulatory protein
|
|
chr10_-_3817418
|
1.265
|
NM_001160124
NM_001160125
NM_001300
|
KLF6
|
Kruppel-like factor 6
|
|
chrX_+_64804318
|
1.264
|
|
MSN
|
moesin
|
|
chrX_+_49084499
|
1.263
|
NM_012196
NM_001472
NM_001127200
NM_001098407
NM_001127212
|
GAGE8
GAGE1
GAGE2C
GAGE2E
GAGE12J
GAGE2D
GAGE2A
|
G antigen 8
G antigen 1
G antigen 2C
G antigen 2E
G antigen 12J
G antigen 2D
G antigen 2A
|
|
chr7_-_150305933
|
1.244
|
NM_000238
NM_172056
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2
|
|
chr7_-_99536966
|
1.240
|
|
MCM7
|
minichromosome maintenance complex component 7
|
|
chrX_+_49065452
|
1.239
|
NM_001098406
|
GAGE1
GAGE4
GAGE5
GAGE6
GAGE7
GAGE12J
|
G antigen 1
G antigen 4
G antigen 5
G antigen 6
G antigen 7
G antigen 12J
|
|
chrX_+_49113081
|
1.235
|
NM_001472
NM_001098407
|
GAGE8
GAGE2C
GAGE7
GAGE2E
GAGE2D
GAGE2A
|
G antigen 8
G antigen 2C
G antigen 7
G antigen 2E
G antigen 2D
G antigen 2A
|
|
chrX_+_49103560
|
1.234
|
NM_012196
NM_001472
NM_001474
NM_001475
NM_021123
NM_001477
NM_001127200
NM_001127212
|
GAGE8
GAGE1
GAGE2C
GAGE4
GAGE5
GAGE6
GAGE7
GAGE12I
GAGE2E
GAGE12B
GAGE2A
|
G antigen 8
G antigen 1
G antigen 2C
G antigen 4
G antigen 5
G antigen 6
G antigen 7
G antigen 12I
G antigen 2E
G antigen 12B
G antigen 2A
|
|
chr7_+_154720547
|
1.224
|
|
INSIG1
|
insulin induced gene 1
|
|
chrX_+_64804259
|
1.216
|
|
MSN
|
moesin
|
|
chr11_-_685010
|
1.216
|
|
DEAF1
|
deformed epidermal autoregulatory factor 1 (Drosophila)
|
|
chr21_-_35343456
|
1.215
|
NM_001754
|
RUNX1
|
runt-related transcription factor 1
|
|
chr11_-_63770983
|
1.214
|
NM_138689
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B
|
|
chrX_+_49183712
|
1.213
|
NM_001127199
NM_001098409
NM_001098408
NM_001127345
NM_001098418
NM_001098410
|
GAGE12D
GAGE1
GAGE4
GAGE5
GAGE6
GAGE7
GAGE12G
GAGE12C
GAGE12B
GAGE12E
GAGE12H
|
G antigen 12D
G antigen 1
G antigen 4
G antigen 5
G antigen 6
G antigen 7
G antigen 12G
G antigen 12C
G antigen 12B
G antigen 12E
G antigen 12H
|
|
chr7_+_149696792
|
1.210
|
NM_001099695
NM_001099696
NM_013400
|
REPIN1
|
replication initiator 1
|
|
chr6_+_36670113
|
1.206
|
|
SRSF3
|
serine/arginine-rich splicing factor 3
|
|
chr12_-_54409067
|
1.205
|
|
CD63
|
CD63 molecule
|