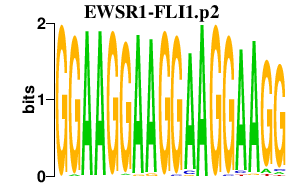
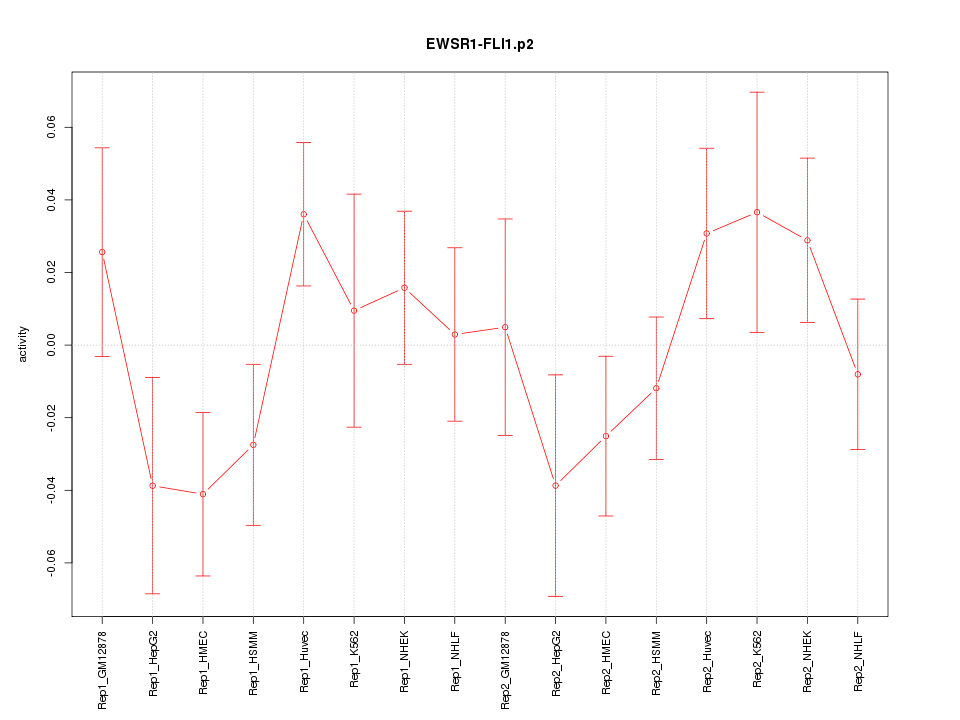
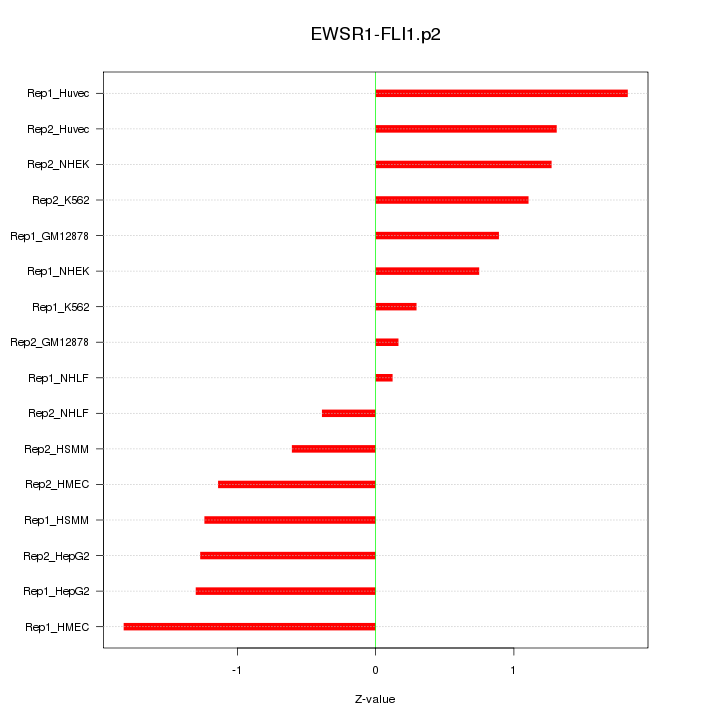
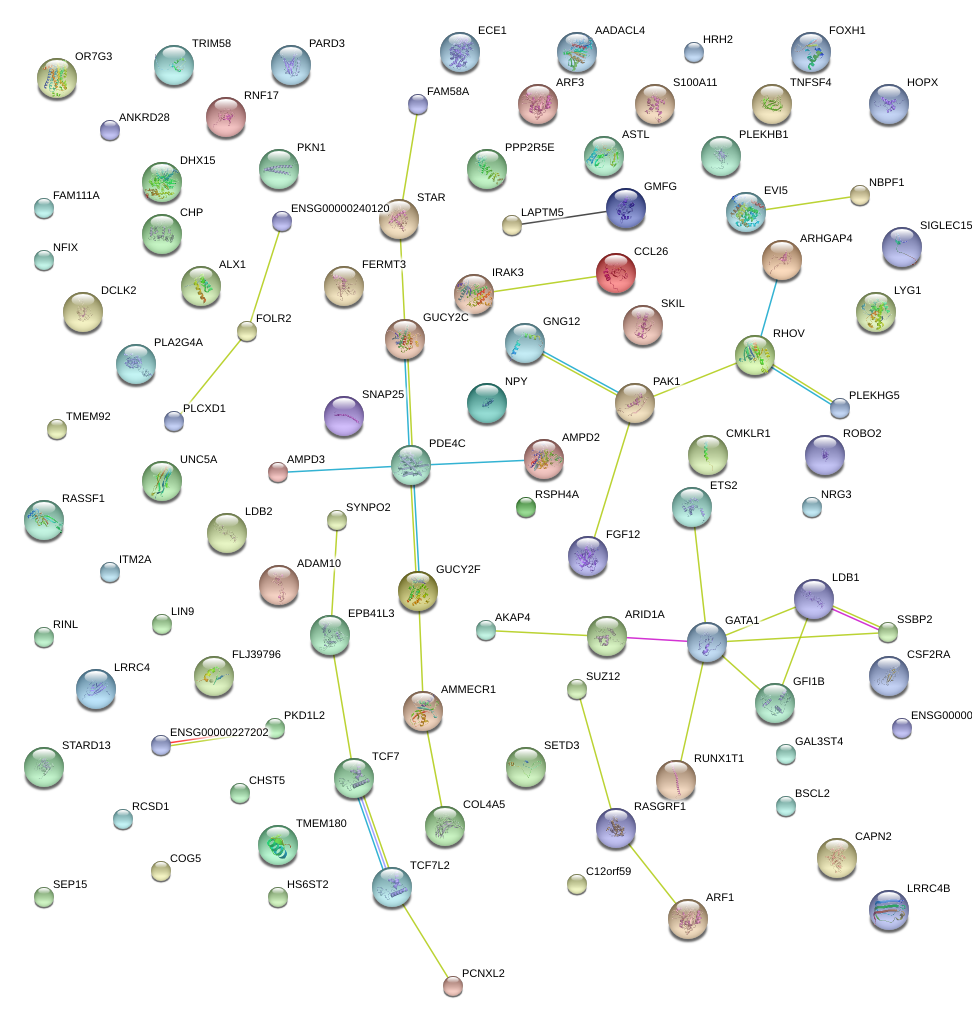
|
chr1_-_93030538
|
3.273
|
NM_005665
|
EVI5
|
ecotropic viral integration site 5
|
|
chr3_-_50353209
|
2.041
|
NM_007182
NM_170714
|
RASSF1
|
Ras association (RalGDS/AF-6) domain family member 1
|
|
chrX_+_1361570
|
1.930
|
NM_001161530
NM_172247
|
CSF2RA
|
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage)
|
|
chr1_+_165865953
|
1.440
|
NM_052862
|
RCSD1
|
RCSD domain containing 1
|
|
chr19_-_55763091
|
1.399
|
NM_001080457
|
LRRC4B
|
leucine rich repeat containing 4B
|
|
chr5_-_71838875
|
1.342
|
NM_152625
|
ZNF366
|
zinc finger protein 366
|
|
chr1_-_31003181
|
1.273
|
|
LAPTM5
|
lysosomal protein transmembrane 5
|
|
chr1_+_246087230
|
1.228
|
|
TRIM58
|
tripartite motif containing 58
|
|
chr8_-_38127489
|
1.173
|
NM_000349
|
STAR
|
steroidogenic acute regulatory protein
|
|
chr1_+_246087104
|
1.157
|
NM_015431
|
TRIM58
|
tripartite motif containing 58
|
|
chr10_+_83627422
|
1.137
|
NM_001165973
|
NRG3
|
neuregulin 3
|
|
chr1_-_31003260
|
1.069
|
NM_006762
|
LAPTM5
|
lysosomal protein transmembrane 5
|
|
chr12_+_84198015
|
1.042
|
NM_006982
|
ALX1
|
ALX homeobox 1
|
|
chr2_-_99284070
|
0.971
|
NM_174898
|
LYG1
|
lysozyme G-like 1
|
|
chrX_-_78509662
|
0.946
|
NM_001171581
NM_004867
|
ITM2A
|
integral membrane protein 2A
|
|
chr1_+_185064654
|
0.941
|
NM_024420
|
PLA2G4A
|
phospholipase A2, group IVA (cytosolic, calcium-dependent)
|
|
chr17_+_45706786
|
0.927
|
NM_153229
|
TMEM92
|
transmembrane protein 92
|
|
chr13_-_32678142
|
0.903
|
NM_052851
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr1_+_165866127
|
0.893
|
|
RCSD1
|
RCSD domain containing 1
|
|
chr11_+_73034870
|
0.825
|
NM_001130033
NM_021200
|
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1
|
|
chr11_+_63730766
|
0.800
|
|
FERMT3
|
fermitin family member 3
|
|
chrX_-_78509367
|
0.794
|
|
ITM2A
|
integral membrane protein 2A
|
|
chr18_-_5620629
|
0.738
|
|
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3
|
|
chr18_+_41659542
|
0.731
|
NM_213602
|
SIGLEC15
|
sialic acid binding Ig-like lectin 15
|
|
chr8_-_93176819
|
0.719
|
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr8_-_145671947
|
0.709
|
|
FOXH1
|
forkhead box H1
|
|
chr20_-_48741481
|
0.700
|
|
|
|
|
chr3_-_193609531
|
0.672
|
NM_021032
|
FGF12
|
fibroblast growth factor 12
|
|
chr11_+_10434056
|
0.653
|
NM_001025390
|
AMPD3
|
adenosine monophosphate deaminase 3
|
|
chr19_-_44060729
|
0.646
|
NM_001195833
NM_198445
|
RINL
|
Ras and Rab interactor-like
|
|
chrX_+_48529905
|
0.623
|
NM_002049
|
GATA1
|
GATA binding protein 1 (globin transcription factor 1)
|
|
chr8_-_93177057
|
0.611
|
NM_001198625
NM_001198626
NM_001198627
NM_001198631
NM_001198634
NM_001198679
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr1_-_231498081
|
0.566
|
NM_014801
|
PCNXL2
|
pecanex-like 2 (Drosophila)
|
|
chr7_-_75256999
|
0.562
|
NM_006072
|
CCL26
|
chemokine (C-C motif) ligand 26
|
|
chr8_-_145672525
|
0.551
|
NM_003923
|
FOXH1
|
forkhead box H1
|
|
chr19_-_9098625
|
0.547
|
NM_001001958
|
OR7G3
|
olfactory receptor, family 7, subfamily G, member 3
|
|
chr12_+_64869271
|
0.547
|
|
IRAK3
|
interleukin-1 receptor-associated kinase 3
|
|
chr12_+_10222823
|
0.545
|
NM_153022
|
C12orf59
|
chromosome 12 open reading frame 59
|
|
chr19_-_4867814
|
0.542
|
|
|
|
|
chrX_+_138060
|
0.536
|
NM_018390
|
PLCXD1
|
phosphatidylinositol-specific phospholipase C, X domain containing 1
|
|
chr11_+_10433241
|
0.531
|
NM_001025389
|
AMPD3
|
adenosine monophosphate deaminase 3
|
|
chr12_+_64869296
|
0.524
|
|
IRAK3
|
interleukin-1 receptor-associated kinase 3
|
|
chr21_+_39099624
|
0.524
|
|
ETS2
|
v-ets erythroblastosis virus E26 oncogene homolog 2 (avian)
|
|
chr4_-_57216816
|
0.506
|
|
HOPX
|
HOP homeobox
|
|
chr15_-_77085056
|
0.483
|
NM_153815
|
RASGRF1
|
Ras protein-specific guanine nucleotide-releasing factor 1
|
|
chr7_-_99604127
|
0.481
|
|
GAL3ST4
|
galactose-3-O-sulfotransferase 4
|
|
chr1_+_12627152
|
0.476
|
NM_001013630
|
AADACL4
|
arylacetamide deacetylase-like 4
|
|
chr7_-_127458231
|
0.474
|
NM_022143
|
LRRC4
|
leucine rich repeat containing 4
|
|
chr19_+_13066956
|
0.472
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr4_-_16509354
|
0.467
|
|
LDB2
|
LIM domain binding 2
|
|
chr1_-_171442937
|
0.465
|
NM_003326
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4
|
|
chr4_-_16509126
|
0.463
|
|
LDB2
|
LIM domain binding 2
|
|
chr6_+_117044334
|
0.453
|
NM_001010892
NM_001161664
|
RSPH4A
|
radial spoke head 4 homolog A (Chlamydomonas)
|
|
chr11_+_63730720
|
0.440
|
NM_031471
NM_178443
|
FERMT3
|
fermitin family member 3
|
|
chrX_-_49851743
|
0.435
|
NM_139289
|
AKAP4
|
A kinase (PRKA) anchor protein 4
|
|
chrX_+_107569671
|
0.431
|
NM_000495
NM_033380
|
COL4A5
|
collagen, type IV, alpha 5
|
|
chr7_+_24290331
|
0.422
|
NM_000905
|
NPY
|
neuropeptide Y
|
|
chr10_+_114700775
|
0.418
|
|
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box)
|
|
chr4_-_16509446
|
0.418
|
NM_001130834
NM_001290
|
LDB2
|
LIM domain binding 2
|
|
chr1_-_21478571
|
0.401
|
NM_001113347
|
ECE1
|
endothelin converting enzyme 1
|
|
chr12_+_64869181
|
0.393
|
NM_001142523
NM_007199
|
IRAK3
|
interleukin-1 receptor-associated kinase 3
|
|
chr9_+_134843918
|
0.393
|
NM_001135031
NM_004188
|
GFI1B
|
growth factor independent 1B transcription repressor
|
|
chr16_-_79811475
|
0.389
|
NM_001076780
NM_052892
|
PKD1L2
|
polycystic kidney disease 1-like 2
|
|
chrX_-_131918905
|
0.380
|
|
HS6ST2
|
heparan sulfate 6-O-sulfotransferase 2
|
|
chrX_-_109447970
|
0.377
|
NM_001025580
NM_015365
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1
|
|
chr7_-_99604171
|
0.376
|
NM_024637
|
GAL3ST4
|
galactose-3-O-sulfotransferase 4
|
|
chr19_-_18220009
|
0.374
|
NM_000923
|
PDE4C
|
phosphodiesterase 4C, cAMP-specific
|
|
chr19_-_44518465
|
0.361
|
|
GMFG
|
glia maturation factor, gamma
|
|
chr11_-_76800525
|
0.361
|
|
PAK1
|
p21 protein (Cdc42/Rac)-activated kinase 1
|
|
chr12_-_92489652
|
0.359
|
|
LOC144481
|
hypothetical protein LOC144481
|
|
chr14_-_63079492
|
0.358
|
|
PPP2R5E
|
protein phosphatase 2, regulatory subunit B', epsilon isoform
|
|
chr14_-_63079299
|
0.355
|
|
PPP2R5E
|
protein phosphatase 2, regulatory subunit B', epsilon isoform
|
|
chr21_+_39099717
|
0.354
|
NM_005239
|
ETS2
|
v-ets erythroblastosis virus E26 oncogene homolog 2 (avian)
|
|
chr16_-_74126568
|
0.353
|
NM_024533
|
CHST5
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 5
|
|
chr3_+_77171938
|
0.352
|
NM_002942
|
ROBO2
|
roundabout, axon guidance receptor, homolog 2 (Drosophila)
|
|
chr15_-_56829069
|
0.342
|
|
ADAM10
|
ADAM metallopeptidase domain 10
|
|
chr5_+_175041069
|
0.336
|
NM_022304
|
HRH2
|
histamine receptor H2
|
|
chr1_-_87152596
|
0.334
|
NM_004261
NM_203341
|
SEP15
|
15 kDa selenoprotein
|
|
chr17_+_27288253
|
0.334
|
|
SUZ12
|
suppressor of zeste 12 homolog (Drosophila)
|
|
chr1_+_109969571
|
0.323
|
NM_203404
|
AMPD2
|
adenosine monophosphate deaminase 2
|
|
chr5_-_64100176
|
0.319
|
|
SREK1IP1
|
SREK1-interacting protein 1
|
|
chr4_-_57217226
|
0.316
|
NM_001145459
NM_139211
|
HOPX
|
HOP homeobox
|
|
chr11_+_58668827
|
0.315
|
NM_022074
NM_198847
|
FAM111A
|
family with sequence similarity 111, member A
|
|
chr1_-_6468600
|
0.310
|
NM_001042665
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5
|
|
chr10_-_103864651
|
0.307
|
NM_003893
|
LDB1
|
LIM domain binding 1
|
|
chr19_-_44518516
|
0.307
|
NM_004877
|
GMFG
|
glia maturation factor, gamma
|
|
chr3_-_15875932
|
0.302
|
NM_015199
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr1_+_221955876
|
0.302
|
NM_001146068
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr5_-_64100239
|
0.301
|
NM_173829
|
SREK1IP1
|
SREK1-interacting protein 1
|
|
chrX_-_109447954
|
0.301
|
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1
|
|
chr1_+_26895108
|
0.295
|
NM_006015
NM_139135
|
ARID1A
|
AT rich interactive domain 1A (SWI-like)
|
|
chr10_+_104211134
|
0.291
|
|
TMEM180
|
transmembrane protein 180
|
|
chr2_-_96167900
|
0.290
|
NM_001002036
|
ASTL
|
astacin-like metallo-endopeptidase (M12 family)
|
|
chr7_-_106991941
|
0.285
|
NM_001161520
NM_006348
NM_181733
|
COG5
|
component of oligomeric golgi complex 5
|
|
chr19_+_54621499
|
0.282
|
NM_001195256
|
LOC645971
|
hypothetical protein LOC645971
|
|
chr11_-_62231358
|
0.279
|
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin)
|
|
chr1_-_68071635
|
0.275
|
NM_018841
|
GNG12
|
guanine nucleotide binding protein (G protein), gamma 12
|
|
chr10_+_104211148
|
0.274
|
NM_024789
|
TMEM180
|
transmembrane protein 180
|
|
chr5_-_81082699
|
0.274
|
NM_012446
|
SSBP2
|
single-stranded DNA binding protein 2
|
|
chr3_+_171558215
|
0.273
|
|
SKIL
|
SKI-like oncogene
|
|
chr3_+_171558188
|
0.270
|
|
SKIL
|
SKI-like oncogene
|
|
chr4_+_120029426
|
0.267
|
NM_001128933
NM_001128934
NM_133477
|
SYNPO2
|
synaptopodin 2
|
|
chr12_-_47637396
|
0.267
|
|
ARF3
|
ADP-ribosylation factor 3
|
|
chr8_-_93176618
|
0.261
|
NM_001198629
NM_001198630
NM_001198632
NM_175635
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr11_+_71605466
|
0.258
|
NM_000803
NM_001113534
NM_001113535
NM_001113536
|
FOLR2
|
folate receptor 2 (fetal)
|
|
chrX_-_109448083
|
0.256
|
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1
|
|
chr10_-_35143900
|
0.253
|
NM_001184785
NM_001184786
NM_001184787
NM_001184788
NM_001184789
NM_001184790
NM_001184791
NM_001184792
NM_001184793
NM_001184794
NM_019619
|
PARD3
|
par-3 partitioning defective 3 homolog (C. elegans)
|
|
chr1_-_150276064
|
0.252
|
|
S100A11
|
S100 calcium binding protein A11
|
|
chr20_+_10147455
|
0.247
|
NM_003081
NM_130811
|
SNAP25
|
synaptosomal-associated protein, 25kDa
|
|
chr15_+_39310757
|
0.246
|
|
CHP
|
calcium binding protein P22
|
|
chrX_-_152844880
|
0.246
|
|
ARHGAP4
|
Rho GTPase activating protein 4
|
|
chr4_+_151218862
|
0.244
|
NM_001040260
NM_001040261
|
DCLK2
|
doublecortin-like kinase 2
|
|
chr1_-_224563461
|
0.241
|
|
LIN9
|
lin-9 homolog (C. elegans)
|
|
chrX_-_108611940
|
0.238
|
NM_001522
|
GUCY2F
|
guanylate cyclase 2F, retinal
|
|
chr12_-_107238568
|
0.237
|
NM_001142345
|
CMKLR1
|
chemokine-like receptor 1
|
|
chr15_-_56829104
|
0.237
|
|
ADAM10
|
ADAM metallopeptidase domain 10
|
|
chr14_-_99016833
|
0.235
|
|
SETD3
|
SET domain containing 3
|
|
chr5_+_137701855
|
0.234
|
|
FAM53C
|
family with sequence similarity 53, member C
|
|
chr5_-_38631263
|
0.233
|
NM_002310
|
LIFR
|
leukemia inhibitory factor receptor alpha
|
|
chr12_-_54869511
|
0.230
|
|
SMARCC2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2
|
|
chr19_-_51820133
|
0.229
|
NM_000960
|
PTGIR
|
prostaglandin I2 (prostacyclin) receptor (IP)
|
|
chr1_+_242691295
|
0.227
|
NM_001130957
NM_173807
|
C1orf101
|
chromosome 1 open reading frame 101
|
|
chr15_+_39310693
|
0.227
|
NM_007236
|
CHP
|
calcium binding protein P22
|
|
chr1_-_109957195
|
0.223
|
NM_005272
|
GNAT2
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2
|
|
chr17_+_27288152
|
0.222
|
NM_015355
|
SUZ12
|
suppressor of zeste 12 homolog (Drosophila)
|
|
chr7_+_128142661
|
0.222
|
NM_032599
|
FAM71F1
|
family with sequence similarity 71, member F1
|
|
chr11_+_2279932
|
0.221
|
|
TSPAN32
|
tetraspanin 32
|
|
chr1_-_150276114
|
0.221
|
NM_005620
|
S100A11
|
S100 calcium binding protein A11
|
|
chr14_+_59782222
|
0.219
|
NM_177952
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A
|
|
chr7_+_21434818
|
0.219
|
|
|
|
|
chr15_+_89248423
|
0.218
|
NM_006122
|
MAN2A2
|
mannosidase, alpha, class 2A, member 2
|
|
chr12_-_47637530
|
0.214
|
|
|
|
|
chr14_-_63079724
|
0.212
|
|
PPP2R5E
|
protein phosphatase 2, regulatory subunit B', epsilon isoform
|
|
chr1_-_150276091
|
0.212
|
|
S100A11
|
S100 calcium binding protein A11
|
|
chr15_-_66285427
|
0.212
|
NM_001031733
NM_033429
|
CALML4
|
calmodulin-like 4
|
|
chr1_+_87153025
|
0.208
|
|
HS2ST1
|
heparan sulfate 2-O-sulfotransferase 1
|
|
chr1_+_160868932
|
0.208
|
|
DDR2
|
discoidin domain receptor tyrosine kinase 2
|
|
chr5_+_137701852
|
0.208
|
|
FAM53C
|
family with sequence similarity 53, member C
|
|
chr14_-_63079816
|
0.207
|
NM_006246
|
PPP2R5E
|
protein phosphatase 2, regulatory subunit B', epsilon isoform
|
|
chr12_-_47637564
|
0.203
|
|
ARF3
|
ADP-ribosylation factor 3
|
|
chr3_-_129852345
|
0.201
|
|
RPN1
|
ribophorin I
|
|
chr1_+_157441133
|
0.201
|
NM_002036
|
DARC
|
Duffy blood group, chemokine receptor
|
|
chr1_+_160868822
|
0.195
|
NM_001014796
NM_006182
|
DDR2
|
discoidin domain receptor tyrosine kinase 2
|
|
chr12_-_47637507
|
0.193
|
|
ARF3
|
ADP-ribosylation factor 3
|
|
chr17_-_8004671
|
0.193
|
|
VAMP2
|
vesicle-associated membrane protein 2 (synaptobrevin 2)
|
|
chr10_-_103864579
|
0.193
|
|
LDB1
|
LIM domain binding 1
|
|
chr19_-_57592778
|
0.191
|
|
LOC284373
|
hypothetical protein LOC284373
|
|
chr17_+_2443652
|
0.188
|
NM_000430
|
PAFAH1B1
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1 (45kDa)
|
|
chr3_-_129852400
|
0.187
|
NM_002950
|
RPN1
|
ribophorin I
|
|
chr12_-_47637518
|
0.185
|
NM_001659
|
ARF3
|
ADP-ribosylation factor 3
|
|
chr12_-_47637512
|
0.184
|
|
ARF3
|
ADP-ribosylation factor 3
|
|
chr11_-_62231445
|
0.183
|
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin)
|
|
chr8_-_37943143
|
0.182
|
NM_000025
|
ADRB3
|
adrenergic, beta-3-, receptor
|
|
chr11_+_10480891
|
0.182
|
|
AMPD3
|
adenosine monophosphate deaminase 3
|
|
chr5_+_137701601
|
0.181
|
NM_016605
|
FAM53C
|
family with sequence similarity 53, member C
|
|
chr11_+_2279812
|
0.180
|
NM_139022
|
TSPAN32
|
tetraspanin 32
|
|
chr12_+_105501161
|
0.180
|
NM_213594
|
RFX4
|
regulatory factor X, 4 (influences HLA class II expression)
|
|
chrX_-_102870207
|
0.178
|
NM_001024452
NM_001172285
|
GLRA4
|
glycine receptor, alpha 4
|
|
chr11_-_62231636
|
0.178
|
NM_001122955
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin)
|
|
chr11_-_62231386
|
0.177
|
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin)
|
|
chr18_+_75388522
|
0.177
|
|
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1
|
|
chr17_+_27288167
|
0.176
|
|
SUZ12
|
suppressor of zeste 12 homolog (Drosophila)
|
|
chr12_+_52060191
|
0.175
|
NM_138473
|
SP1
|
Sp1 transcription factor
|
|
chr2_+_113533155
|
0.172
|
NM_012275
|
IL1F5
|
interleukin 1 family, member 5 (delta)
|
|
chr19_+_57592913
|
0.169
|
NM_032423
|
ZNF528
|
zinc finger protein 528
|
|
chrX_+_24077649
|
0.168
|
NM_001178086
NM_001178095
NM_001178084
NM_001178085
NM_003410
|
ZFX
|
zinc finger protein, X-linked
|
|
chr17_+_27288178
|
0.167
|
|
SUZ12
|
suppressor of zeste 12 homolog (Drosophila)
|
|
chr7_+_49783774
|
0.165
|
NM_198570
|
VWC2
|
von Willebrand factor C domain containing 2
|
|
chr1_+_28936030
|
0.164
|
NM_001172828
|
YTHDF2
|
YTH domain family, member 2
|
|
chr11_-_67163600
|
0.162
|
NM_005995
|
TBX10
|
T-box 10
|
|
chr11_-_2279642
|
0.161
|
NM_001142946
|
C11orf21
|
chromosome 11 open reading frame 21
|
|
chr1_+_157441824
|
0.161
|
NM_001122951
|
DARC
|
Duffy blood group, chemokine receptor
|
|
chr1_-_184684478
|
0.159
|
NM_022576
|
PDC
|
phosducin
|
|
chr3_-_193928038
|
0.159
|
NM_004113
|
FGF12
|
fibroblast growth factor 12
|
|
chr18_+_54682261
|
0.159
|
|
ZNF532
|
zinc finger protein 532
|
|
chrX_+_56606686
|
0.158
|
NM_013444
|
UBQLN2
|
ubiquilin 2
|
|
chr5_+_150020595
|
0.156
|
NM_001122853
NM_133371
|
MYOZ3
|
myozenin 3
|
|
chr17_+_24598000
|
0.156
|
NM_005208
|
CRYBA1
|
crystallin, beta A1
|
|
chrX_+_67635348
|
0.156
|
NM_001195214
NM_173834
|
YIPF6
|
Yip1 domain family, member 6
|
|
chr8_-_133562185
|
0.156
|
NM_004519
|
KCNQ3
|
potassium voltage-gated channel, KQT-like subfamily, member 3
|
|
chr17_+_27288374
|
0.155
|
|
|
|
|
chr9_-_138701721
|
0.154
|
NM_001012727
NM_006412
|
AGPAT2
|
1-acylglycerol-3-phosphate O-acyltransferase 2 (lysophosphatidic acid acyltransferase, beta)
|
|
chr4_-_96689137
|
0.154
|
|
UNC5C
|
unc-5 homolog C (C. elegans)
|
|
chr11_-_117628211
|
0.153
|
NM_198275
|
MPZL3
|
myelin protein zero-like 3
|
|
chr15_+_81906983
|
0.152
|
NM_003027
|
SH3GL3
|
SH3-domain GRB2-like 3
|
|
chr9_+_35528628
|
0.151
|
NM_001135999
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr6_-_130578110
|
0.149
|
NM_152552
|
SAMD3
|
sterile alpha motif domain containing 3
|
|
chr1_+_153241665
|
0.148
|
NM_015872
|
ZBTB7B
|
zinc finger and BTB domain containing 7B
|
|
chr7_+_128218685
|
0.147
|
|
CCDC136
|
coiled-coil domain containing 136
|
|
chr5_+_156819782
|
0.146
|
|
NIPAL4
|
NIPA-like domain containing 4
|
|
chr1_-_155936904
|
0.146
|
NM_052939
|
FCRL3
|
Fc receptor-like 3
|
|
chr9_+_35528890
|
0.144
|
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr12_+_88627601
|
0.144
|
|
LOC338758
|
hypothetical LOC338758
|
|
chr21_+_38550533
|
0.144
|
NM_002243
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15
|
|
chr1_+_44185005
|
0.143
|
NM_014652
|
IPO13
|
importin 13
|
|
chr14_-_63079840
|
0.142
|
|
PPP2R5E
|
protein phosphatase 2, regulatory subunit B', epsilon isoform
|
|
chr19_+_54883732
|
0.142
|
NM_001101340
|
C19orf76
|
chromosome 19 open reading frame 76
|
|
chr12_-_40824699
|
0.142
|
NM_001099650
NM_173601
|
GXYLT1
|
glucoside xylosyltransferase 1
|
|
chr1_+_87152905
|
0.139
|
NM_001134492
NM_012262
|
HS2ST1
|
heparan sulfate 2-O-sulfotransferase 1
|
|
chr2_+_182464910
|
0.137
|
|
SSFA2
|
sperm specific antigen 2
|