|
chr7_+_106292975
|
8.069
|
|
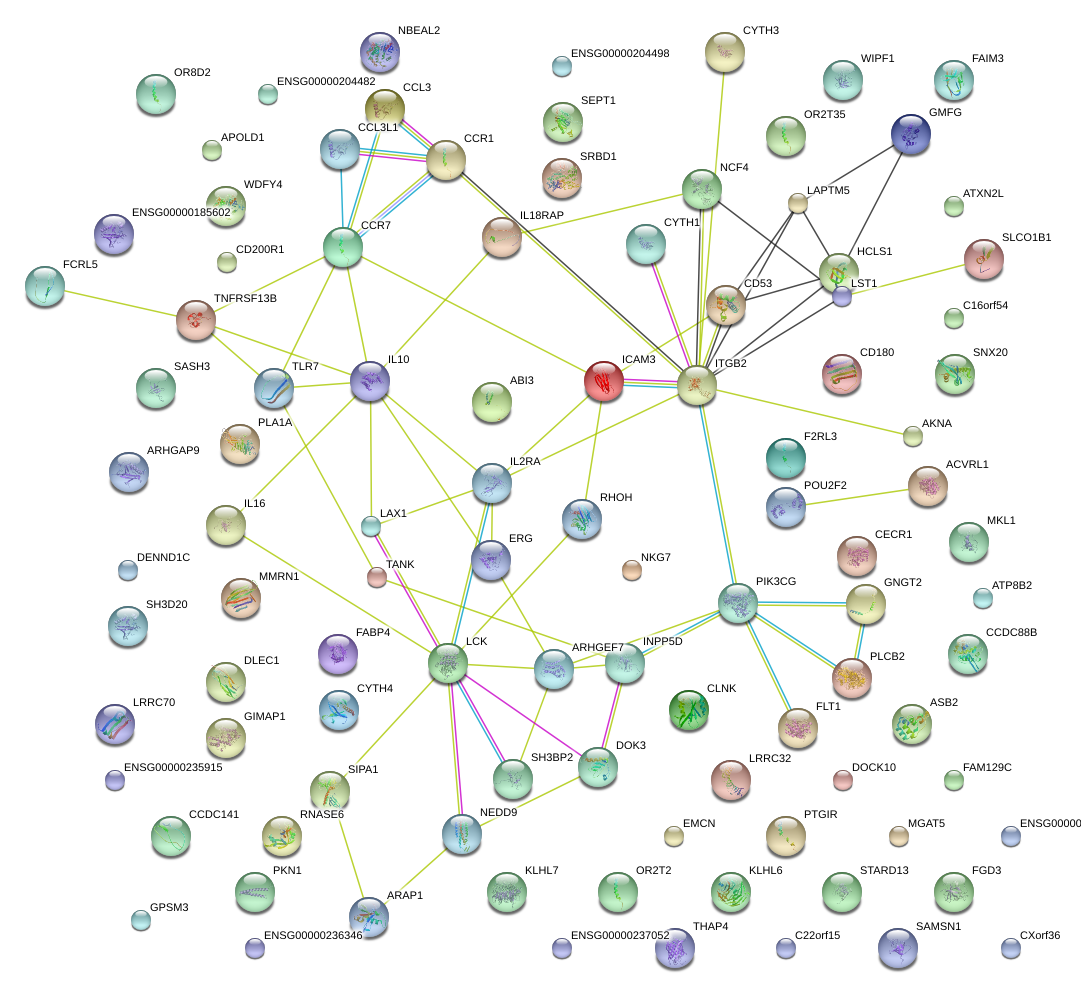
PIK3CG
|
phosphoinositide-3-kinase, catalytic, gamma polypeptide
|
|
chr7_+_106292932
|
8.003
|
|
PIK3CG
|
phosphoinositide-3-kinase, catalytic, gamma polypeptide
|
|
chr5_+_61910318
|
7.209
|
NM_181506
|
LRRC70
|
leucine rich repeat containing 70
|
|
chr3_-_184756101
|
7.050
|
NM_130446
|
KLHL6
|
kelch-like 6 (Drosophila)
|
|
chr3_-_122862391
|
6.262
|
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr11_+_65164167
|
6.191
|
NM_153253
|
SIPA1
|
signal-induced proliferation-associated 1
|
|
chr7_+_106293112
|
6.104
|
NM_002649
|
PIK3CG
|
phosphoinositide-3-kinase, catalytic, gamma polypeptide
|
|
chr3_-_122862447
|
6.076
|
NM_005335
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr13_+_110653407
|
5.285
|
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7
|
|
chr17_-_31441390
|
4.878
|
NM_002983
|
CCL3
|
chemokine (C-C motif) ligand 3
|
|
chr10_+_49562866
|
4.851
|
|
WDFY4
|
WDFY family member 4
|
|
chr2_-_175207521
|
4.757
|
NM_003387
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr15_+_79376295
|
4.691
|
NM_004513
|
IL16
|
interleukin 16 (lymphocyte chemoattractant factor)
|
|
chr12_-_56157898
|
4.598
|
NM_001080156
|
ARHGAP9
|
Rho GTPase activating protein 9
|
|
chr3_+_120799411
|
4.556
|
NM_015900
|
PLA1A
|
phospholipase A1 member A
|
|
chr14_+_20319049
|
4.266
|
NM_005615
|
RNASE6
|
ribonuclease, RNase A family, k6
|
|
chr1_-_155788775
|
4.242
|
NM_001195388
NM_031281
|
FCRL5
|
Fc receptor-like 5
|
|
chr3_-_46224791
|
4.071
|
NM_001295
|
CCR1
|
chemokine (C-C motif) receptor 1
|
|
chr6_-_11490487
|
3.981
|
NM_001142393
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr8_-_82557956
|
3.808
|
NM_001442
|
FABP4
|
fatty acid binding protein 4, adipocyte
|
|
chr4_+_39874921
|
3.797
|
NM_004310
|
RHOH
|
ras homolog gene family, member H
|
|
chr17_-_40843530
|
3.780
|
|
ARHGAP27
|
Rho GTPase activating protein 27
|
|
chr3_-_13032130
|
3.725
|
|
|
|
|
chr12_-_56159631
|
3.724
|
NM_001080157
|
ARHGAP9
|
Rho GTPase activating protein 9
|
|
chr19_-_56567602
|
3.723
|
NM_005601
|
NKG7
|
natural killer cell group 7 sequence
|
|
chr22_-_39189369
|
3.714
|
|
MKL1
|
megakaryoblastic leukemia (translocation) 1
|
|
chr4_+_91035025
|
3.699
|
NM_007351
|
MMRN1
|
multimerin 1
|
|
chr3_-_114176401
|
3.687
|
NM_138806
NM_138939
NM_138940
NM_170780
|
CD200R1
|
CD200 receptor 1
|
|
chr12_-_56159895
|
3.653
|
NM_032496
|
ARHGAP9
|
Rho GTPase activating protein 9
|
|
chr6_-_32268267
|
3.593
|
|
GPSM3
|
G-protein signaling modulator 3
|
|
chr2_-_225519974
|
3.592
|
|
DOCK10
|
dedicator of cytokinesis 10
|
|
chr2_+_134728299
|
3.577
|
NM_002410
|
MGAT5
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase
|
|
chr19_-_51820133
|
3.557
|
NM_000960
|
PTGIR
|
prostaglandin I2 (prostacyclin) receptor (IP)
|
|
chr4_-_101658094
|
3.546
|
NM_001159694
NM_016242
|
EMCN
|
endomucin
|
|
chr22_+_35586950
|
3.499
|
NM_000631
NM_013416
|
NCF4
|
neutrophil cytosolic factor 4, 40kDa
|
|
chr21_-_14840506
|
3.481
|
NM_022136
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1
|
|
chr19_+_16860871
|
3.438
|
|
F2RL3
|
coagulation factor II (thrombin) receptor-like 3
|
|
chr19_-_10311262
|
3.429
|
NM_002162
|
ICAM3
|
intercellular adhesion molecule 3
|
|
chr4_+_2783743
|
3.425
|
NM_001145855
|
SH3BP2
|
SH3-domain binding protein 2
|
|
chr4_+_2783764
|
3.382
|
|
SH3BP2
|
SH3-domain binding protein 2
|
|
chr11_+_65164767
|
3.366
|
|
SIPA1
|
signal-induced proliferation-associated 1
|
|
chr17_-_16816113
|
3.357
|
NM_012452
|
TNFRSF13B
|
tumor necrosis factor receptor superfamily, member 13B
|
|
chr19_+_16860807
|
3.351
|
NM_003950
|
F2RL3
|
coagulation factor II (thrombin) receptor-like 3
|
|
chr1_-_205012348
|
3.324
|
NM_000572
|
IL10
|
interleukin 10
|
|
chr9_-_116190038
|
3.232
|
|
AKNA
|
AT-hook transcription factor
|
|
chr2_+_161701711
|
3.215
|
NM_004180
NM_133484
|
TANK
|
TRAF family member-associated NFKB activator
|
|
chr13_-_27967215
|
3.094
|
NM_001159920
NM_001160030
NM_001160031
NM_002019
|
FLT1
|
fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor)
|
|
chr22_-_16080265
|
3.058
|
|
CECR1
|
cat eye syndrome chromosome region, candidate 1
|
|
chr1_-_246869181
|
3.020
|
NM_001001827
|
OR2T35
|
olfactory receptor, family 2, subfamily T, member 35
|
|
chr19_+_17495109
|
3.000
|
NM_001098524
NM_173544
|
FAM129C
|
family with sequence similarity 129, member C
|
|
chr1_+_202000906
|
2.984
|
NM_001136190
NM_017773
|
LAX1
|
lymphocyte transmembrane adaptor 1
|
|
chr19_-_44518465
|
2.973
|
|
GMFG
|
glia maturation factor, gamma
|
|
chr3_+_47004457
|
2.960
|
|
NBEAL2
|
neurobeachin-like 2
|
|
chr6_-_32268615
|
2.959
|
|
GPSM3
|
G-protein signaling modulator 3
|
|
chr13_-_32658215
|
2.917
|
NM_178007
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr16_-_29664765
|
2.903
|
NM_175900
|
C16orf54
|
chromosome 16 open reading frame 54
|
|
chr6_+_31661934
|
2.865
|
NM_205838
NM_205839
|
LST1
|
leukocyte specific transcript 1
|
|
chr11_+_63864263
|
2.849
|
NM_032251
|
CCDC88B
|
coiled-coil domain containing 88B
|
|
chr1_-_205161821
|
2.840
|
NM_001193338
NM_001142473
NM_005449
|
FAIM3
|
Fas apoptotic inhibitory molecule 3
|
|
chr2_-_45648608
|
2.717
|
|
SRBD1
|
S1 RNA binding domain 1
|
|
chr2_-_242205556
|
2.716
|
NM_001164356
|
THAP4
|
THAP domain containing 4
|
|
chr5_-_66528330
|
2.701
|
NM_005582
|
CD180
|
CD180 molecule
|
|
chr15_-_75259714
|
2.663
|
|
PEAK1
|
NKF3 kinase family member
|
|
chr4_-_10295412
|
2.652
|
NM_052964
|
CLNK
|
cytokine-dependent hematopoietic cell linker
|
|
chr11_-_72111027
|
2.648
|
NM_001135190
NM_015242
|
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1
|
|
chr21_-_38792173
|
2.608
|
NM_001136155
NM_182918
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr16_-_49272617
|
2.606
|
|
SNX20
|
sorting nexin 20
|
|
chr1_-_111544603
|
2.598
|
|
|
|
|
chr19_-_6432765
|
2.587
|
NM_024898
|
DENND1C
|
DENN/MADD domain containing 1C
|
|
chr17_+_44642587
|
2.587
|
NM_001135186
NM_016428
|
ABI3
|
ABI family, member 3
|
|
chr1_+_152566899
|
2.583
|
NM_020452
|
ATP8B2
|
ATPase, class I, type 8B, member 2
|
|
chr1_+_111544469
|
2.570
|
|
|
|
|
chr2_+_233633348
|
2.527
|
|
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa
|
|
chr11_-_76059435
|
2.521
|
NM_001128922
|
LRRC32
|
leucine rich repeat containing 32
|
|
chr12_+_12829807
|
2.502
|
NM_030817
|
APOLD1
|
apolipoprotein L domain containing 1
|
|
chrX_+_128741562
|
2.467
|
NM_018990
|
SASH3
|
SAM and SH3 domain containing 3
|
|
chr2_+_233633279
|
2.443
|
NM_001017915
NM_005541
|
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa
|
|
chr17_-_44642934
|
2.442
|
NM_001198754
|
GNGT2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2
|
|
chr2_-_179623030
|
2.433
|
NM_173648
|
CCDC141
|
coiled-coil domain containing 141
|
|
chr5_-_176869943
|
2.431
|
NM_001144875
NM_001144876
|
DOK3
|
docking protein 3
|
|
chr1_+_111217244
|
2.405
|
NM_000560
|
CD53
|
CD53 molecule
|
|
chr19_-_44518516
|
2.397
|
NM_004877
|
GMFG
|
glia maturation factor, gamma
|
|
chr22_+_36008511
|
2.373
|
|
CYTH4
|
cytohesin 4
|
|
chr14_-_93512818
|
2.337
|
|
ASB2
|
ankyrin repeat and SOCS box containing 2
|
|
chr12_+_50592379
|
2.335
|
NM_001077401
|
ACVRL1
|
activin A receptor type II-like 1
|
|
chr16_-_30301357
|
2.321
|
|
SEPT1
|
septin 1
|
|
chr16_-_49272741
|
2.313
|
NM_001144972
NM_153337
NM_182854
|
SNX20
|
sorting nexin 20
|
|
chr17_-_35975227
|
2.312
|
NM_001838
|
CCR7
|
chemokine (C-C motif) receptor 7
|
|
chr10_+_49563520
|
2.299
|
NM_020945
|
WDFY4
|
WDFY family member 4
|
|
chr21_-_45165204
|
2.294
|
|
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit)
|
|
chr2_+_102401580
|
2.293
|
NM_003853
|
IL18RAP
|
interleukin 18 receptor accessory protein
|
|
chr9_+_94766379
|
2.293
|
|
FGD3
|
FYVE, RhoGEF and PH domain containing 3
|
|
chr3_+_38055699
|
2.292
|
NM_007335
NM_007337
|
DLEC1
|
deleted in lung and esophageal cancer 1
|
|
chrX_+_128741578
|
2.289
|
|
SASH3
|
SAM and SH3 domain containing 3
|
|
chrX_+_12795111
|
2.287
|
NM_016562
|
TLR7
|
toll-like receptor 7
|
|
chr22_+_22435207
|
2.273
|
NM_182520
|
C22orf15
|
chromosome 22 open reading frame 15
|
|
chr10_-_6144119
|
2.270
|
NM_000417
|
IL2RA
|
interleukin 2 receptor, alpha
|
|
chr19_-_47328399
|
2.257
|
NM_002698
|
POU2F2
|
POU class 2 homeobox 2
|
|
chr1_+_111217314
|
2.253
|
|
CD53
|
CD53 molecule
|
|
chr21_-_45165249
|
2.243
|
|
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit)
|
|
chr19_+_14412087
|
2.231
|
|
PKN1
|
protein kinase N1
|
|
chrX_-_44945025
|
2.216
|
NM_024689
NM_176819
|
CXorf36
|
chromosome X open reading frame 36
|
|
chr1_-_31003260
|
2.202
|
NM_006762
|
LAPTM5
|
lysosomal protein transmembrane 5
|
|
chr15_-_38387164
|
2.174
|
|
PLCB2
|
phospholipase C, beta 2
|
|
chr1_+_32489426
|
2.174
|
NM_005356
|
LCK
|
lymphocyte-specific protein tyrosine kinase
|
|
chr7_+_150044530
|
2.172
|
NM_130759
|
GIMAP1
|
GTPase, IMAP family member 1
|
|
chr7_-_3049872
|
2.164
|
NM_032415
|
CARD11
|
caspase recruitment domain family, member 11
|
|
chr22_+_36008465
|
2.158
|
|
CYTH4
|
cytohesin 4
|
|
chr1_-_79244948
|
2.155
|
NM_022159
|
ELTD1
|
EGF, latrophilin and seven transmembrane domain containing 1
|
|
chr17_+_4283967
|
2.145
|
NM_182538
|
SPNS3
|
spinster homolog 3 (Drosophila)
|
|
chr22_+_36008369
|
2.126
|
NM_013385
|
CYTH4
|
cytohesin 4
|
|
chr10_+_26767657
|
2.109
|
|
APBB1IP
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein
|
|
chr1_+_157067729
|
2.066
|
NM_002432
|
MNDA
|
myeloid cell nuclear differentiation antigen
|
|
chr3_-_150533948
|
2.061
|
NM_001184723
NM_138786
|
TM4SF18
|
transmembrane 4 L six family member 18
|
|
chr1_-_158815850
|
2.051
|
|
CD84
|
CD84 molecule
|
|
chr1_+_207995999
|
2.042
|
NM_025228
|
TRAF3IP3
|
TRAF3 interacting protein 3
|
|
chr11_+_63730720
|
2.041
|
NM_031471
NM_178443
|
FERMT3
|
fermitin family member 3
|
|
chr16_+_56134037
|
2.040
|
|
GPR114
|
G protein-coupled receptor 114
|
|
chr7_-_44985175
|
2.037
|
|
MYO1G
|
myosin IG
|
|
chr12_+_9033403
|
2.028
|
NM_005810
|
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1
|
|
chr5_-_156468614
|
2.022
|
NM_032782
|
HAVCR2
|
hepatitis A virus cellular receptor 2
|
|
chr14_-_93512647
|
2.019
|
|
ASB2
|
ankyrin repeat and SOCS box containing 2
|
|
chr1_-_181826292
|
1.969
|
NM_000433
NM_001190789
NM_001190794
|
NCF2
|
neutrophil cytosolic factor 2
|
|
chr16_+_56134077
|
1.968
|
NM_153837
|
GPR114
|
G protein-coupled receptor 114
|
|
chr7_-_44985202
|
1.957
|
NM_033054
|
MYO1G
|
myosin IG
|
|
chr11_-_72057755
|
1.947
|
NM_001146209
|
PDE2A
|
phosphodiesterase 2A, cGMP-stimulated
|
|
chr9_-_129656413
|
1.945
|
|
ENG
|
endoglin
|
|
chrX_-_130792297
|
1.943
|
|
LOC286467
|
hypothetical LOC286467
|
|
chrX_-_47374187
|
1.941
|
NM_001145252
|
CFP
|
complement factor properdin
|
|
chr11_+_63730766
|
1.930
|
|
FERMT3
|
fermitin family member 3
|
|
chr10_+_26767698
|
1.928
|
|
APBB1IP
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein
|
|
chr1_-_158815895
|
1.928
|
NM_001184879
NM_001184881
NM_001184882
NM_003874
|
CD84
|
CD84 molecule
|
|
chr9_+_263025
|
1.921
|
NM_001190458
NM_001193536
|
DOCK8
|
dedicator of cytokinesis 8
|
|
chr16_+_30391609
|
1.917
|
|
|
|
|
chr1_+_170077234
|
1.914
|
NM_001136127
NM_015569
|
DNM3
|
dynamin 3
|
|
chr1_+_196874867
|
1.913
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr11_+_1830763
|
1.901
|
NM_002339
|
LSP1
|
lymphocyte-specific protein 1
|
|
chr1_-_159306037
|
1.891
|
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr1_-_155834357
|
1.888
|
NM_031282
|
FCRL4
|
Fc receptor-like 4
|
|
chr15_-_38387266
|
1.885
|
|
PLCB2
|
phospholipase C, beta 2
|
|
chr19_-_4510713
|
1.876
|
|
SEMA6B
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6B
|
|
chr19_-_1083109
|
1.845
|
NM_001100122
|
SBNO2
|
strawberry notch homolog 2 (Drosophila)
|
|
chr1_+_196874839
|
1.842
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr16_+_30391550
|
1.835
|
|
ITGAL
|
integrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide)
|
|
chr12_+_25096472
|
1.826
|
NM_006152
|
LRMP
|
lymphoid-restricted membrane protein
|
|
chr11_-_110755112
|
1.821
|
|
POU2AF1
|
POU class 2 associating factor 1
|
|
chr7_+_69869099
|
1.818
|
|
AUTS2
|
autism susceptibility candidate 2
|
|
chr16_-_30301634
|
1.816
|
NM_052838
|
SEPT1
|
septin 1
|
|
chr2_+_33514896
|
1.788
|
NM_170672
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated)
|
|
chr22_-_35970230
|
1.785
|
NM_002872
|
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2)
|
|
chr14_+_92188546
|
1.783
|
|
RIN3
|
Ras and Rab interactor 3
|
|
chr15_-_38387317
|
1.782
|
|
PLCB2
|
phospholipase C, beta 2
|
|
chr21_-_45165302
|
1.769
|
NM_000211
|
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit)
|
|
chr7_+_149842850
|
1.768
|
NM_153236
|
GIMAP7
|
GTPase, IMAP family member 7
|
|
chr5_-_156468571
|
1.766
|
|
HAVCR2
|
hepatitis A virus cellular receptor 2
|
|
chr11_+_22644732
|
1.766
|
NM_001143830
|
GAS2
|
growth arrest-specific 2
|
|
chr7_-_149960612
|
1.759
|
NM_024711
|
GIMAP6
|
GTPase, IMAP family member 6
|
|
chr17_-_1495742
|
1.757
|
NM_003693
NM_145350
NM_145352
|
SCARF1
|
scavenger receptor class F, member 1
|
|
chr9_+_107464558
|
1.755
|
NM_005421
|
TAL2
|
T-cell acute lymphocytic leukemia 2
|
|
chr1_-_159306210
|
1.748
|
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr6_-_154719560
|
1.739
|
NM_001130700
NM_015553
|
IPCEF1
|
interaction protein for cytohesin exchange factors 1
|
|
chr16_+_30391460
|
1.730
|
NM_001114380
NM_002209
|
ITGAL
|
integrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide)
|
|
chr10_-_88707299
|
1.729
|
NM_024756
|
MMRN2
|
multimerin 2
|
|
chr7_-_44985084
|
1.724
|
|
MYO1G
|
myosin IG
|
|
chr17_-_18890963
|
1.717
|
NM_006613
|
GRAP
|
GRB2-related adaptor protein
|
|
chr15_-_38387357
|
1.716
|
|
PLCB2
|
phospholipase C, beta 2
|
|
chr2_+_130703384
|
1.701
|
|
|
|
|
chr1_+_196874759
|
1.701
|
NM_002838
NM_080921
NM_080923
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr12_-_56422120
|
1.700
|
NM_014770
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2
|
|
chr6_+_31662454
|
1.694
|
NM_205837
|
LST1
|
leukocyte specific transcript 1
|
|
chr17_+_4922478
|
1.691
|
|
ZFP3
|
zinc finger protein 3 homolog (mouse)
|
|
chr22_-_35970133
|
1.686
|
|
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2)
|
|
chr7_+_91762227
|
1.679
|
|
|
|
|
chr15_-_41879426
|
1.675
|
NM_001033517
|
SERINC4
|
serine incorporator 4
|
|
chr15_-_38387410
|
1.673
|
NM_004573
|
PLCB2
|
phospholipase C, beta 2
|
|
chr17_-_26672757
|
1.671
|
NM_001003927
NM_014210
|
EVI2A
|
ecotropic viral integration site 2A
|
|
chr9_-_129656581
|
1.670
|
|
ENG
|
endoglin
|
|
chr12_-_107745455
|
1.666
|
NM_001161331
|
SSH1
|
slingshot homolog 1 (Drosophila)
|
|
chr5_-_169657192
|
1.655
|
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr6_+_167456230
|
1.655
|
NM_031409
|
CCR6
|
chemokine (C-C motif) receptor 6
|
|
chrX_-_50403412
|
1.654
|
|
SHROOM4
|
shroom family member 4
|
|
chr1_-_159306273
|
1.649
|
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr2_-_136590210
|
1.645
|
NM_001008540
|
CXCR4
|
chemokine (C-X-C motif) receptor 4
|
|
chr1_-_181826086
|
1.644
|
|
NCF2
|
neutrophil cytosolic factor 2
|
|
chr1_+_111818111
|
1.643
|
NM_174896
|
C1orf162
|
chromosome 1 open reading frame 162
|
|
chr19_+_10820105
|
1.643
|
NM_001136482
|
C19orf38
|
chromosome 19 open reading frame 38
|
|
chr1_-_111544784
|
1.642
|
NM_024901
|
DENND2D
|
DENN/MADD domain containing 2D
|
|
chr15_+_42815851
|
1.641
|
NM_080745
NM_182985
|
TRIM69
|
tripartite motif containing 69
|
|
chr9_-_129707392
|
1.638
|
|
LOC100131355
ST6GALNAC6
|
hypothetical protein LOC100131355
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6
|
|
chr2_+_182029863
|
1.632
|
NM_000885
|
ITGA4
|
integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor)
|
|
chr15_+_89229275
|
1.631
|
NM_001143783
NM_001143784
|
FES
|
feline sarcoma oncogene
|
|
chr5_+_143171918
|
1.630
|
NM_021182
|
HMHB1
|
histocompatibility (minor) HB-1
|
|
chr17_+_4922445
|
1.627
|
NM_153018
|
ZFP3
|
zinc finger protein 3 homolog (mouse)
|
|
chr11_-_110755366
|
1.627
|
NM_006235
|
POU2AF1
|
POU class 2 associating factor 1
|
|
chr15_-_78050515
|
1.624
|
NM_001114735
NM_004049
|
BCL2A1
|
BCL2-related protein A1
|
|
chr5_-_149515613
|
1.622
|
NM_002609
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr8_+_75066117
|
1.619
|
NM_001195797
NM_015364
|
LY96
|
lymphocyte antigen 96
|
|
chr12_+_6751930
|
1.617
|
NM_002286
|
LAG3
|
lymphocyte-activation gene 3
|
|
chr9_-_35744271
|
1.616
|
NM_001044264
|
MSMP
|
microseminoprotein, prostate associated
|
|
chr9_-_70780574
|
1.605
|
|
|
|