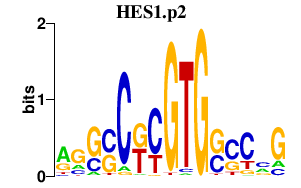
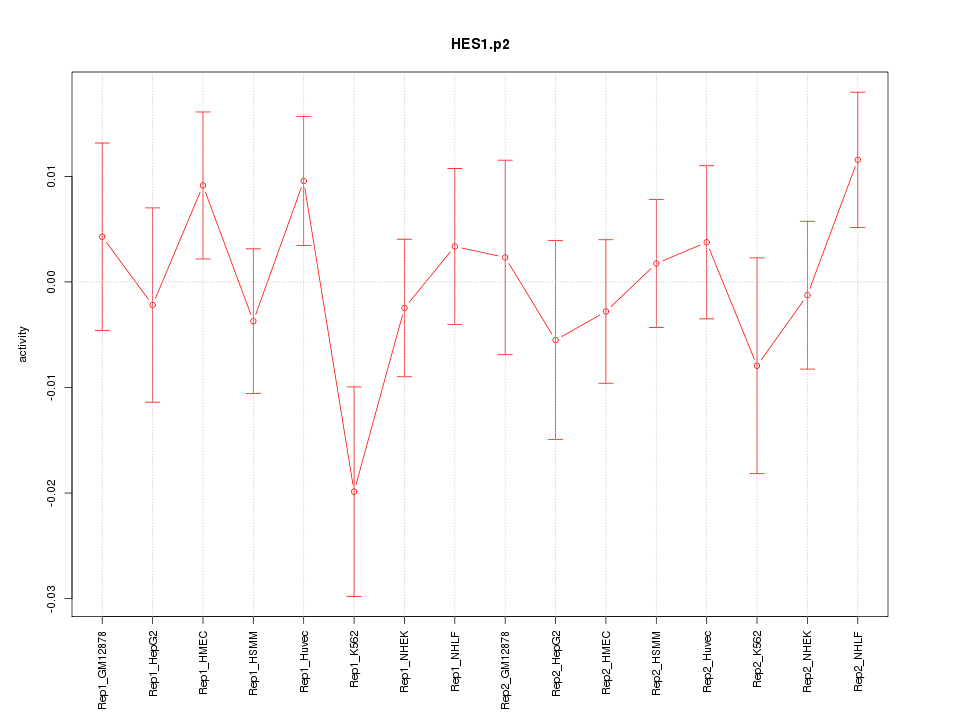
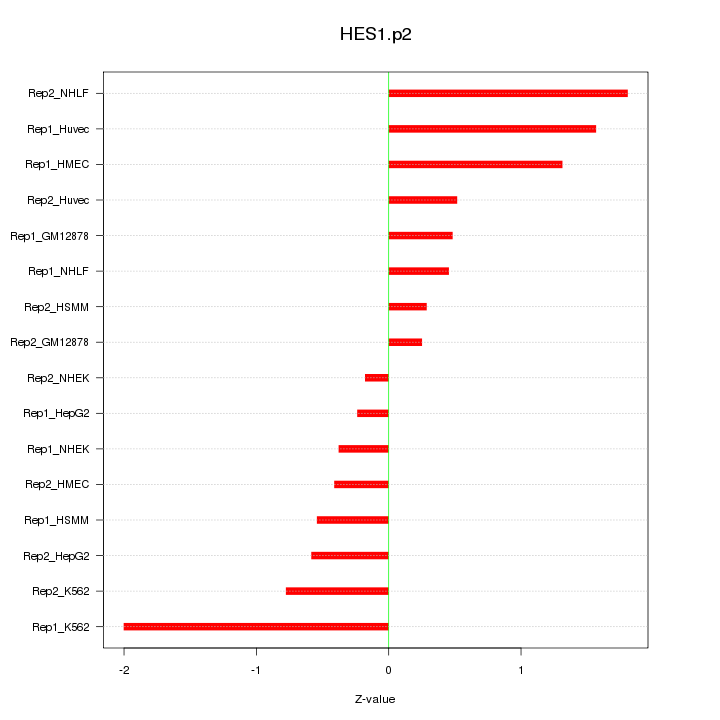
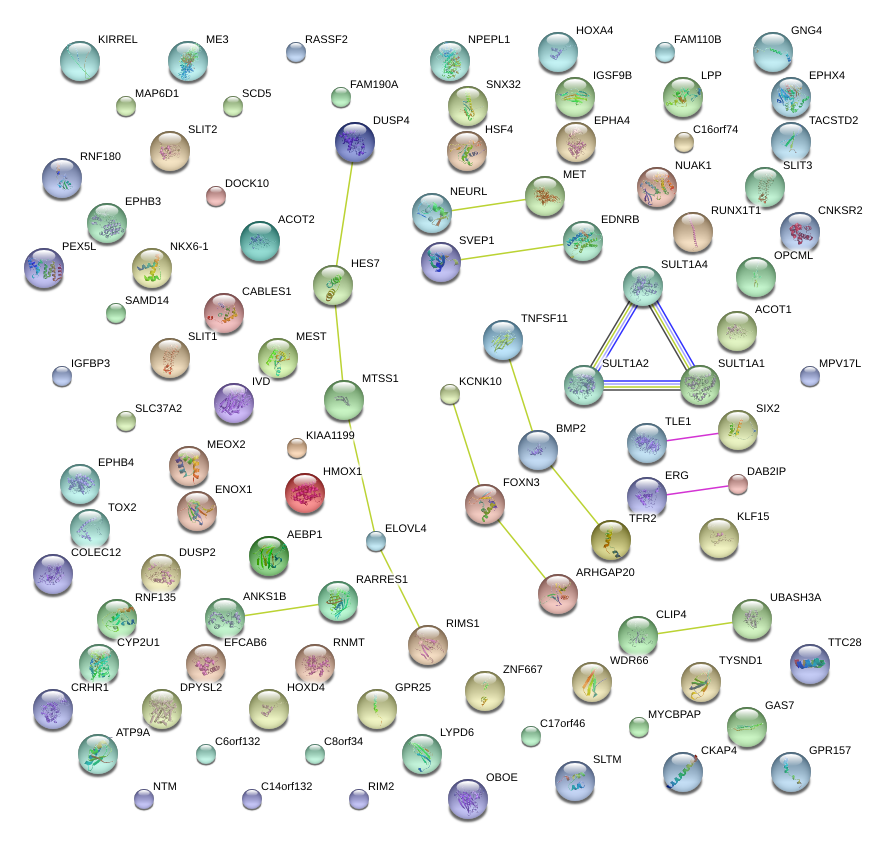
|
chr2_-_225615556
|
2.021
|
NM_014689
|
DOCK10
|
dedicator of cytokinesis 10
|
|
chr2_+_149895274
|
1.321
|
NM_194317
|
LYPD6
|
LY6/PLAUR domain containing 6
|
|
chr14_+_73073570
|
1.278
|
NM_001037161
|
ACOT1
|
acyl-CoA thioesterase 1
|
|
chr6_+_72949092
|
1.252
|
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr1_+_199108705
|
1.040
|
NM_005298
|
GPR25
|
G protein-coupled receptor 25
|
|
chr17_-_10042579
|
1.036
|
NM_201433
|
GAS7
|
growth arrest-specific 7
|
|
chr1_-_9111674
|
1.031
|
NM_024980
|
GPR157
|
G protein-coupled receptor 157
|
|
chr8_+_69405510
|
0.996
|
NM_001195639
NM_052958
|
C8orf34
|
chromosome 8 open reading frame 34
|
|
chr18_+_18969724
|
0.845
|
NM_001100619
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1
|
|
chr13_-_77390907
|
0.837
|
NM_001122659
NM_003991
|
EDNRB
|
endothelin receptor type B
|
|
chr17_-_76985538
|
0.819
|
|
|
|
|
chr3_-_189354510
|
0.809
|
|
LOC339929
|
hypothetical LOC339929
|
|
chr21_-_38954433
|
0.802
|
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr9_+_123501186
|
0.767
|
|
DAB2IP
|
DAB2 interacting protein
|
|
chr19_-_61680510
|
0.767
|
NM_022103
|
ZNF667
|
zinc finger protein 667
|
|
chr14_+_73105618
|
0.750
|
|
ACOT2
|
acyl-CoA thioesterase 2
|
|
chr7_-_15692551
|
0.738
|
|
MEOX2
|
mesenchyme homeobox 2
|
|
chr14_-_87859284
|
0.736
|
NM_138317
|
KCNK10
|
potassium channel, subfamily K, member 10
|
|
chr3_-_185025989
|
0.730
|
NM_024871
|
MAP6D1
|
MAP6 domain containing 1
|
|
chr4_+_19864332
|
0.723
|
NM_004787
|
SLIT2
|
slit homolog 2 (Drosophila)
|
|
chr16_-_28542366
|
0.721
|
NM_177536
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1
|
|
chr11_-_86061003
|
0.716
|
NM_006680
|
ME3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial
|
|
chrX_+_21302317
|
0.714
|
NM_001168647
NM_001168648
NM_001168649
NM_014927
|
CNKSR2
|
connector enhancer of kinase suppressor of Ras 2
|
|
chr12_+_120840829
|
0.713
|
NM_001178003
NM_144668
|
WDR66
|
WD repeat domain 66
|
|
chr11_-_86060812
|
0.710
|
NM_001014811
|
ME3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial
|
|
chr20_-_49818306
|
0.685
|
NM_006045
|
ATP9A
|
ATPase, class II, type 9A
|
|
chr8_+_59069522
|
0.663
|
NM_147189
|
FAM110B
|
family with sequence similarity 110, member B
|
|
chr9_-_112381593
|
0.659
|
NM_153366
|
SVEP1
|
sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1
|
|
chr3_+_189354167
|
0.655
|
NM_001167672
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr22_+_34107087
|
0.646
|
|
HMOX1
|
heme oxygenase (decycling) 1
|
|
chr6_-_80713589
|
0.642
|
NM_022726
|
ELOVL4
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 4
|
|
chr22_+_34107051
|
0.629
|
NM_002133
|
HMOX1
|
heme oxygenase (decycling) 1
|
|
chr7_-_100068612
|
0.627
|
|
TFR2
|
transferrin receptor 2
|
|
chr15_+_38485310
|
0.621
|
|
IVD
|
isovaleryl-CoA dehydrogenase
|
|
chr7_-_45927263
|
0.612
|
NM_000598
NM_001013398
|
IGFBP3
|
insulin-like growth factor binding protein 3
|
|
chr11_+_65357972
|
0.612
|
NM_152760
|
SNX32
|
sorting nexin 32
|
|
chr12_-_105057940
|
0.612
|
NM_014840
|
NUAK1
|
NUAK family, SNF1-like kinase, 1
|
|
chr20_-_49818268
|
0.608
|
|
ATP9A
|
ATPase, class II, type 9A
|
|
chr7_+_129918408
|
0.599
|
NM_177525
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr2_-_96174865
|
0.599
|
NM_004418
|
DUSP2
|
dual specificity phosphatase 2
|
|
chr13_-_43259032
|
0.596
|
NM_017993
|
ENOX1
|
ecto-NOX disulfide-thiol exchanger 1
|
|
chr4_+_91267706
|
0.594
|
NM_001145065
|
FAM190A
|
family with sequence similarity 190, member A
|
|
chr17_-_40695216
|
0.575
|
NM_152343
|
C17orf46
|
chromosome 17 open reading frame 46
|
|
chr11_+_131285747
|
0.573
|
NM_001144058
NM_001144059
NM_016522
|
NTM
|
neurotrimin
|
|
chr5_+_63497408
|
0.569
|
NM_001113561
NM_178532
|
RNF180
|
ring finger protein 180
|
|
chr2_-_96174502
|
0.563
|
|
DUSP2
|
dual specificity phosphatase 2
|
|
chr17_+_45940911
|
0.556
|
|
MYCBPAP
|
MYCBP associated protein
|
|
chr8_+_26427378
|
0.546
|
NM_001197293
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr11_+_124438407
|
0.544
|
|
SLC37A2
|
solute carrier family 37 (glycerol-3-phosphate transporter), member 2
|
|
chr7_-_27136876
|
0.542
|
NM_002141
|
HOXA4
|
homeobox A4
|
|
chr17_-_7968121
|
0.532
|
NM_001165967
NM_032580
|
HES7
|
hairy and enhancer of split 7 (Drosophila)
|
|
chr18_-_490583
|
0.529
|
NM_130386
|
COLEC12
|
collectin sub-family member 12
|
|
chr20_-_4752067
|
0.525
|
NM_014737
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2
|
|
chr4_-_83938920
|
0.524
|
NM_001037582
NM_024906
|
SCD5
|
stearoyl-CoA desaturase 5
|
|
chr4_+_109072120
|
0.520
|
NM_183075
|
CYP2U1
|
cytochrome P450, family 2, subfamily U, polypeptide 1
|
|
chr15_+_78858763
|
0.514
|
NM_018689
|
KIAA1199
|
KIAA1199
|
|
chr1_-_58815753
|
0.513
|
NM_002353
|
TACSTD2
|
tumor-associated calcium signal transducer 2
|
|
chr17_+_45940743
|
0.508
|
NM_032133
|
MYCBPAP
|
MYCBP associated protein
|
|
chr1_-_233879617
|
0.507
|
NM_001098722
|
GNG4
|
guanine nucleotide binding protein (G protein), gamma 4
|
|
chr20_+_41978195
|
0.493
|
NM_001098796
NM_032883
|
TOX2
|
TOX high mobility group box family member 2
|
|
chr9_-_21549696
|
0.491
|
|
LOC554202
|
hypothetical LOC554202
|
|
chr13_+_42046182
|
0.490
|
NM_003701
|
TNFSF11
|
tumor necrosis factor (ligand) superfamily, member 11
|
|
chr11_-_110087982
|
0.487
|
|
ARHGAP20
|
Rho GTPase activating protein 20
|
|
chr12_-_98902562
|
0.484
|
NM_152788
|
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B
|
|
chr20_+_6696744
|
0.483
|
NM_001200
|
BMP2
|
bone morphogenetic protein 2
|
|
chr10_-_88720908
|
0.482
|
|
LOC100133190
|
hypothetical LOC100133190
|
|
chr14_-_88953059
|
0.481
|
NM_005197
|
FOXN3
|
forkhead box N3
|
|
chr16_-_84342129
|
0.480
|
NM_206967
|
C16orf74
|
chromosome 16 open reading frame 74
|
|
chr8_-_93184629
|
0.478
|
NM_001198633
NM_175634
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr22_-_42539399
|
0.478
|
NM_022785
NM_198856
|
EFCAB6
|
EF-hand calcium binding domain 6
|
|
chr11_-_133332089
|
0.476
|
NM_014987
|
IGSF9B
|
immunoglobulin superfamily, member 9B
|
|
chr7_+_44110527
|
0.475
|
|
AEBP1
|
AE binding protein 1
|
|
chr2_+_29191711
|
0.474
|
NM_024692
|
CLIP4
|
CAP-GLY domain containing linker protein family, member 4
|
|
chr12_-_105165805
|
0.470
|
NM_006825
|
CKAP4
|
cytoskeleton-associated protein 4
|
|
chr7_+_116099648
|
0.470
|
NM_000245
NM_001127500
|
MET
|
met proto-oncogene (hepatocyte growth factor receptor)
|
|
chr3_-_159932965
|
0.468
|
NM_002888
NM_206963
|
RARRES1
|
retinoic acid receptor responder (tazarotene induced) 1
|
|
chr14_-_73106243
|
0.464
|
|
|
|
|
chr16_+_65754788
|
0.461
|
NM_001040667
NM_001538
|
HSF4
|
heat shock transcription factor 4
|
|
chr1_+_156229934
|
0.459
|
|
KIRREL
|
kin of IRRE like (Drosophila)
|
|
chr10_+_105334704
|
0.459
|
|
NEURL
|
neuralized homolog (Drosophila)
|
|
chr3_-_127558747
|
0.458
|
NM_014079
|
KLF15
|
Kruppel-like factor 15
|
|
chr3_-_159932899
|
0.457
|
|
RARRES1
|
retinoic acid receptor responder (tazarotene induced) 1
|
|
chr15_+_38485292
|
0.455
|
|
IVD
|
isovaleryl-CoA dehydrogenase
|
|
chr22_-_27405708
|
0.454
|
NM_001145418
|
TTC28
|
tetratricopeptide repeat domain 28
|
|
chr2_+_177210683
|
0.453
|
|
|
|
|
chr8_-_29262240
|
0.453
|
NM_057158
|
DUSP4
|
dual specificity phosphatase 4
|
|
chr9_-_83493415
|
0.452
|
NM_005077
|
TLE1
|
transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila)
|
|
chr6_-_42218692
|
0.451
|
NM_001164446
|
C6orf132
|
chromosome 6 open reading frame 132
|
|
chr8_-_125809559
|
0.451
|
|
MTSS1
|
metastasis suppressor 1
|
|
chr15_+_38485269
|
0.450
|
|
IVD
|
isovaleryl-CoA dehydrogenase
|
|
chr2_-_45090025
|
0.449
|
NM_016932
|
SIX2
|
SIX homeobox 2
|
|
chr17_+_26322168
|
0.446
|
|
RNF135
|
ring finger protein 135
|
|
chr14_+_95575319
|
0.445
|
|
C14orf132
|
chromosome 14 open reading frame 132
|
|
chr16_+_15397120
|
0.445
|
|
MPV17L
|
MPV17 mitochondrial membrane protein-like
|
|
chr10_-_71575953
|
0.443
|
|
TYSND1
|
trypsin domain containing 1
|
|
chr20_+_56701224
|
0.441
|
NM_024663
|
NPEPL1
|
aminopeptidase-like 1
|
|
chr17_+_26322081
|
0.441
|
NM_001184992
NM_032322
NM_197939
|
RNF135
|
ring finger protein 135
|
|
chr2_-_222145240
|
0.440
|
NM_004438
|
EPHA4
|
EPH receptor A4
|
|
chr3_-_181237210
|
0.440
|
NM_016559
|
PEX5L
|
peroxisomal biogenesis factor 5-like
|
|
chr17_-_45562112
|
0.439
|
NM_174920
|
SAMD14
|
sterile alpha motif domain containing 14
|
|
chr8_+_104582151
|
0.437
|
NM_001100117
|
RIMS2
|
regulating synaptic membrane exocytosis 2
|
|
chr1_+_92268072
|
0.436
|
NM_173567
|
EPHX4
|
epoxide hydrolase 4
|
|
chr3_-_129023850
|
0.435
|
NM_007283
|
MGLL
|
monoglyceride lipase
|
|
chr1_-_45444812
|
0.434
|
NM_020883
|
ZSWIM5
|
zinc finger, SWIM-type containing 5
|
|
chr13_+_20175481
|
0.431
|
NM_138284
|
IL17D
|
interleukin 17D
|
|
chr18_-_75894902
|
0.431
|
|
|
|
|
chr11_+_19691397
|
0.426
|
NM_145117
NM_182964
|
NAV2
|
neuron navigator 2
|
|
chr4_+_2789686
|
0.425
|
NM_001145856
|
SH3BP2
|
SH3-domain binding protein 2
|
|
chr2_-_177210904
|
0.424
|
|
LOC375295
|
hypothetical LOC375295
|
|
chr5_+_175725050
|
0.424
|
NM_173664
|
ARL10
|
ADP-ribosylation factor-like 10
|
|
chr11_+_124438217
|
0.419
|
NM_001145290
NM_198277
|
SLC37A2
|
solute carrier family 37 (glycerol-3-phosphate transporter), member 2
|
|
chr17_-_959007
|
0.418
|
NM_001092
|
ABR
|
active BCR-related gene
|
|
chr18_-_72336133
|
0.417
|
NM_014643
|
ZNF516
|
zinc finger protein 516
|
|
chr6_+_19945578
|
0.415
|
NM_001546
|
ID4
|
inhibitor of DNA binding 4, dominant negative helix-loop-helix protein
|
|
chr2_-_127580975
|
0.415
|
NM_004305
NM_139343
NM_139344
NM_139345
NM_139346
NM_139347
NM_139348
NM_139349
NM_139350
NM_139351
|
BIN1
|
bridging integrator 1
|
|
chr11_-_110088097
|
0.415
|
|
ARHGAP20
|
Rho GTPase activating protein 20
|
|
chr19_-_19599973
|
0.411
|
NM_004720
|
LPAR2
|
lysophosphatidic acid receptor 2
|
|
chr16_-_85099941
|
0.411
|
|
LOC400550
|
hypothetical LOC400550
|
|
chr6_-_112682038
|
0.407
|
|
LAMA4
|
laminin, alpha 4
|
|
chr7_-_47588160
|
0.405
|
|
TNS3
|
tensin 3
|
|
chr16_+_15397087
|
0.404
|
NM_001128423
NM_173803
|
MPV17L
|
MPV17 mitochondrial membrane protein-like
|
|
chr7_+_129919134
|
0.404
|
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr14_+_75114692
|
0.404
|
NM_017791
|
FLVCR2
|
feline leukemia virus subgroup C cellular receptor family, member 2
|
|
chr6_-_150431864
|
0.402
|
NM_024518
|
ULBP3
|
UL16 binding protein 3
|
|
chr8_+_120955080
|
0.401
|
NM_022783
|
DEPTOR
|
DEP domain containing MTOR-interacting protein
|
|
chr11_-_60819110
|
0.401
|
|
VWCE
|
von Willebrand factor C and EGF domains
|
|
chr1_+_234372454
|
0.400
|
NM_003272
|
GPR137B
|
G protein-coupled receptor 137B
|
|
chr18_+_54682261
|
0.397
|
|
ZNF532
|
zinc finger protein 532
|
|
chr13_+_30378311
|
0.393
|
NM_032849
|
C13orf33
|
chromosome 13 open reading frame 33
|
|
chr17_-_829655
|
0.393
|
NM_022463
|
NXN
|
nucleoredoxin
|
|
chr8_-_26427304
|
0.393
|
NM_007257
|
PNMA2
|
paraneoplastic antigen MA2
|
|
chr3_+_106568334
|
0.392
|
NM_001627
|
ALCAM
|
activated leukocyte cell adhesion molecule
|
|
chr2_-_175255717
|
0.391
|
NM_001077269
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr12_-_15833383
|
0.390
|
|
EPS8
|
epidermal growth factor receptor pathway substrate 8
|
|
chr7_-_47588266
|
0.389
|
|
TNS3
|
tensin 3
|
|
chr12_+_15366712
|
0.387
|
NM_002848
NM_030667
|
PTPRO
|
protein tyrosine phosphatase, receptor type, O
|
|
chr2_-_135193011
|
0.384
|
NM_030923
|
TMEM163
|
transmembrane protein 163
|
|
chr13_-_27967215
|
0.384
|
NM_001159920
NM_001160030
NM_001160031
NM_002019
|
FLT1
|
fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor)
|
|
chr17_-_25112393
|
0.382
|
|
SSH2
|
slingshot homolog 2 (Drosophila)
|
|
chr6_-_72187168
|
0.379
|
|
C6orf155
|
chromosome 6 open reading frame 155
|
|
chr3_-_129024665
|
0.378
|
NM_001003794
|
MGLL
|
monoglyceride lipase
|
|
chr13_-_100866839
|
0.377
|
|
NALCN
|
sodium leak channel, non-selective
|
|
chr17_-_53920699
|
0.375
|
NM_001080439
|
HSF5
|
heat shock transcription factor family member 5
|
|
chrX_+_105856531
|
0.375
|
NM_194463
|
RNF128
|
ring finger protein 128
|
|
chr12_-_37585602
|
0.372
|
NM_153634
|
CPNE8
|
copine VIII
|
|
chr6_-_137407504
|
0.371
|
|
IL20RA
|
interleukin 20 receptor, alpha
|
|
chr3_-_49370755
|
0.370
|
NM_000581
NM_201397
|
GPX1
|
glutathione peroxidase 1
|
|
chr1_-_223907283
|
0.369
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr5_+_118719591
|
0.368
|
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8
|
|
chr18_-_42590989
|
0.368
|
NM_013305
|
ST8SIA5
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5
|
|
chr12_-_15833617
|
0.367
|
NM_004447
|
EPS8
|
epidermal growth factor receptor pathway substrate 8
|
|
chr3_+_9826643
|
0.364
|
NM_001025930
|
TTLL3
|
tubulin tyrosine ligase-like family, member 3
|
|
chr2_-_73350103
|
0.363
|
NM_001080410
|
FBXO41
|
F-box protein 41
|
|
chr1_+_2975590
|
0.362
|
NM_022114
NM_199454
|
PRDM16
|
PR domain containing 16
|
|
chr7_+_129919157
|
0.360
|
NM_002402
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr11_-_122571046
|
0.357
|
|
CLMP
|
CXADR-like membrane protein
|
|
chr11_-_62233600
|
0.356
|
NM_001130702
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin)
|
|
chr12_+_121825021
|
0.356
|
NM_032573
NM_201435
|
CCDC62
|
coiled-coil domain containing 62
|
|
chr9_+_17124974
|
0.356
|
NM_001114395
NM_017738
|
CNTLN
|
centlein, centrosomal protein
|
|
chr7_-_15692798
|
0.356
|
NM_005924
|
MEOX2
|
mesenchyme homeobox 2
|
|
chr18_-_53621277
|
0.355
|
NM_005603
|
ATP8B1
|
ATPase, aminophospholipid transporter, class I, type 8B, member 1
|
|
chr3_+_44571600
|
0.354
|
NM_018651
NM_025169
|
ZNF167
|
zinc finger protein 167
|
|
chr4_+_2790338
|
0.351
|
NM_003023
|
SH3BP2
|
SH3-domain binding protein 2
|
|
chrX_-_140098959
|
0.350
|
NM_012317
|
LDOC1
|
leucine zipper, down-regulated in cancer 1
|
|
chr2_+_207016525
|
0.349
|
NM_003812
|
ADAM23
|
ADAM metallopeptidase domain 23
|
|
chr11_-_86061301
|
0.349
|
NM_001161586
|
ME3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial
|
|
chr3_+_112273464
|
0.348
|
NM_015480
|
PVRL3
|
poliovirus receptor-related 3
|
|
chr11_-_122571198
|
0.348
|
NM_024769
|
CLMP
|
CXADR-like membrane protein
|
|
chr5_+_80292213
|
0.347
|
|
RASGRF2
|
Ras protein-specific guanine nucleotide-releasing factor 2
|
|
chr16_-_265868
|
0.347
|
NM_003834
NM_183337
|
RGS11
|
regulator of G-protein signaling 11
|
|
chr6_+_89912464
|
0.346
|
NM_001010853
|
PM20D2
|
peptidase M20 domain containing 2
|
|
chr3_+_54131731
|
0.346
|
NM_018398
|
CACNA2D3
|
calcium channel, voltage-dependent, alpha 2/delta subunit 3
|
|
chr18_+_54681657
|
0.345
|
|
ZNF532
|
zinc finger protein 532
|
|
chr11_-_62233552
|
0.344
|
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin)
|
|
chr11_+_104986620
|
0.338
|
NM_001112812
|
GRIA4
|
glutamate receptor, ionotrophic, AMPA 4
|
|
chr4_-_171247757
|
0.338
|
NM_016228
|
AADAT
|
aminoadipate aminotransferase
|
|
chr15_+_82113813
|
0.338
|
NM_207517
|
ADAMTSL3
|
ADAMTS-like 3
|
|
chr15_-_43457916
|
0.338
|
|
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase)
|
|
chr11_-_88864061
|
0.337
|
NM_001143836
NM_016931
|
NOX4
|
NADPH oxidase 4
|
|
chr6_+_86216457
|
0.337
|
|
NT5E
|
5'-nucleotidase, ecto (CD73)
|
|
chr9_-_112840064
|
0.337
|
NM_001401
NM_057159
|
LPAR1
|
lysophosphatidic acid receptor 1
|
|
chr5_+_118719448
|
0.336
|
NM_014350
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8
|
|
chr7_-_139987044
|
0.336
|
|
DENND2A
|
DENN/MADD domain containing 2A
|
|
chr8_+_28407613
|
0.335
|
NM_017412
|
FZD3
|
frizzled homolog 3 (Drosophila)
|
|
chr11_+_43920385
|
0.331
|
NM_001145033
|
C11orf96
|
chromosome 11 open reading frame 96
|
|
chr1_+_226937732
|
0.330
|
|
RHOU
|
ras homolog gene family, member U
|
|
chr18_+_2836979
|
0.329
|
NM_032048
|
EMILIN2
|
elastin microfibril interfacer 2
|
|
chr10_+_129595289
|
0.329
|
NM_006504
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E
|
|
chr5_-_57791544
|
0.328
|
NM_006622
|
PLK2
|
polo-like kinase 2
|
|
chr13_-_76358433
|
0.328
|
NM_138444
|
KCTD12
|
potassium channel tetramerisation domain containing 12
|
|
chr2_+_9532087
|
0.328
|
NM_001039613
|
IAH1
|
isoamyl acetate-hydrolyzing esterase 1 homolog (S. cerevisiae)
|
|
chrX_-_13866565
|
0.327
|
NM_001001994
|
GPM6B
|
glycoprotein M6B
|
|
chr3_+_54131613
|
0.326
|
|
CACNA2D3
|
calcium channel, voltage-dependent, alpha 2/delta subunit 3
|
|
chr5_+_74197473
|
0.322
|
|
LOC441086
|
hypothetical LOC441086
|
|
chr5_+_110587546
|
0.322
|
NM_001744
|
CAMK4
|
calcium/calmodulin-dependent protein kinase IV
|
|
chr2_-_175060001
|
0.322
|
NM_001033045
NM_152529
|
GPR155
|
G protein-coupled receptor 155
|
|
chr4_+_79197747
|
0.321
|
NM_001166133
NM_025074
|
FRAS1
|
Fraser syndrome 1
|
|
chr7_-_27149750
|
0.320
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chr4_+_169789327
|
0.318
|
NM_001166109
|
PALLD
|
palladin, cytoskeletal associated protein
|
|
chr9_+_71848555
|
0.316
|
|
MAMDC2
|
MAM domain containing 2
|