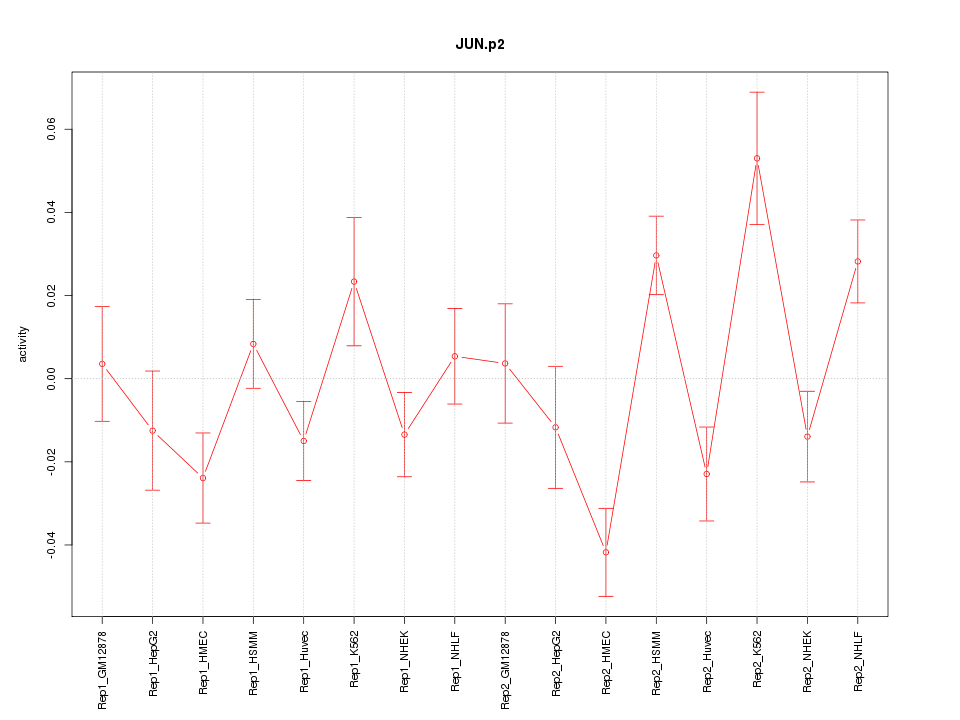
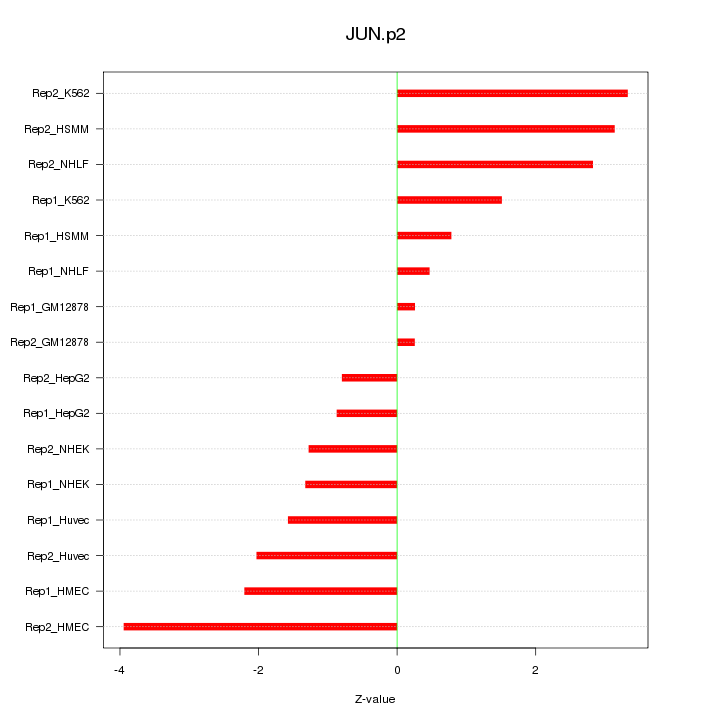
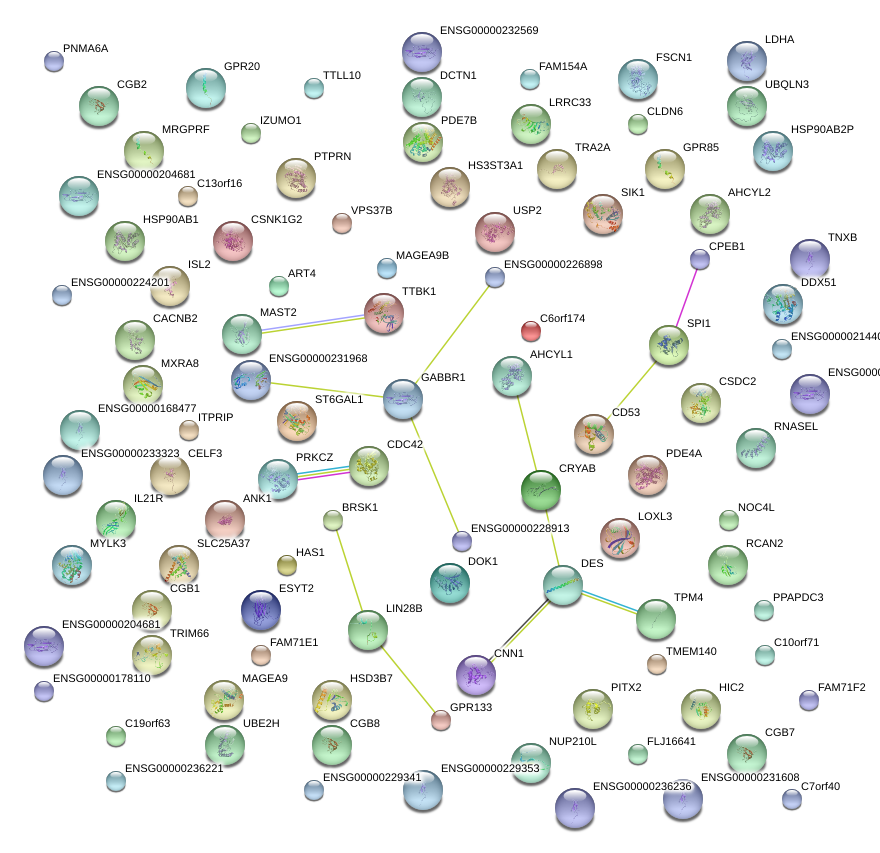
|
chr7_+_134483305
|
2.501
|
NM_018295
|
TMEM140
|
transmembrane protein 140
|
|
chr22_+_40286957
|
2.302
|
NM_014460
|
CSDC2
|
cold shock domain containing C2, RNA binding
|
|
chr2_-_74634569
|
2.173
|
NM_032603
|
LOXL3
|
lysyl oxidase-like 3
|
|
chr19_-_53941835
|
2.160
|
NM_182575
|
IZUMO1
|
izumo sperm-egg fusion 1
|
|
chr11_-_118740069
|
2.114
|
NM_171997
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr1_-_1283770
|
2.071
|
NM_032348
|
MXRA8
|
matrix-remodelling associated 8
|
|
chr15_+_74416156
|
2.059
|
NM_145805
|
ISL2
|
ISL LIM homeobox 2
|
|
chr2_+_74635354
|
1.959
|
|
DOK1
|
docking protein 1, 62kDa (downstream of tyrosine kinase 1)
|
|
chr3_+_188222337
|
1.945
|
NM_003032
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1
|
|
chr11_-_118739838
|
1.896
|
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr15_+_74416129
|
1.884
|
|
ISL2
|
ISL LIM homeobox 2
|
|
chr6_-_127822227
|
1.862
|
NM_014702
|
KIAA0408
|
KIAA0408
|
|
chr9_-_19023183
|
1.845
|
NM_153707
|
FAM154A
|
family with sequence similarity 154, member A
|
|
chr9_+_133154896
|
1.763
|
NM_032728
|
PPAPDC3
|
phosphatidic acid phosphatase type 2 domain containing 3
|
|
chr19_-_56919032
|
1.665
|
NM_001523
|
HAS1
|
hyaluronan synthase 1
|
|
chr8_-_41641881
|
1.659
|
NM_001142445
NM_020478
NM_020480
|
ANK1
|
ankyrin 1, erythrocytic
|
|
chr16_+_30903963
|
1.643
|
NM_001142777
NM_001142778
NM_025193
|
HSD3B7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7
|
|
chr7_-_44992729
|
1.583
|
|
C7orf40
|
chromosome 7 open reading frame 40
|
|
chr12_-_121946561
|
1.551
|
|
|
|
|
chr2_+_219991342
|
1.528
|
NM_001927
|
DES
|
desmin
|
|
chr19_-_55671273
|
1.527
|
|
FAM71E1
|
family with sequence similarity 71, member E1
|
|
chr10_+_50177192
|
1.525
|
NM_001135196
NM_199459
|
C10orf71
|
chromosome 10 open reading frame 71
|
|
chr19_+_16048266
|
1.501
|
|
TPM4
|
tropomyosin 4
|
|
chr1_+_111217314
|
1.478
|
|
CD53
|
CD53 molecule
|
|
chr19_+_11510578
|
1.473
|
NM_001299
|
CNN1
|
calponin 1, basic, smooth muscle
|
|
chr11_-_5487728
|
1.439
|
NM_017481
|
UBQLN3
|
ubiquilin 3
|
|
chr6_+_43319199
|
1.433
|
NM_032538
|
TTBK1
|
tau tubulin kinase 1
|
|
chr8_-_142446546
|
1.373
|
NM_005293
|
GPR20
|
G protein-coupled receptor 20
|
|
chr16_-_3008162
|
1.360
|
NM_021195
|
CLDN6
|
claudin 6
|
|
chr3_+_158026769
|
1.360
|
NM_001004316
NM_001193283
|
LEKR1
|
leucine, glutamate and lysine rich 1
|
|
chr12_+_131194967
|
1.358
|
|
NOC4L
|
nucleolar complex associated 4 homolog (S. cerevisiae)
|
|
chr11_-_68537243
|
1.354
|
NM_001098515
NM_145015
|
MRGPRF
|
MAS-related GPR, member F
|
|
chr16_+_27320983
|
1.347
|
NM_181078
NM_181079
|
IL21R
|
interleukin 21 receptor
|
|
chr22_+_20101661
|
1.345
|
NM_015094
|
HIC2
|
hypermethylated in cancer 2
|
|
chr19_+_55671468
|
1.339
|
NM_175063
NM_206538
|
C19orf63
|
chromosome 19 open reading frame 63
|
|
chr19_+_16048306
|
1.322
|
|
TPM4
|
tropomyosin 4
|
|
chr19_+_54226941
|
1.309
|
NM_033378
|
CGB1
CGB2
|
chorionic gonadotropin, beta polypeptide 1
chorionic gonadotropin, beta polypeptide 2
|
|
chr6_-_166321516
|
1.281
|
|
C6orf176
|
chromosome 6 open reading frame 176
|
|
chr12_+_130004404
|
1.272
|
NM_198827
|
GPR133
|
G protein-coupled receptor 133
|
|
chr10_-_106083652
|
1.237
|
NM_033397
|
ITPRIP
|
inositol 1,4,5-triphosphate receptor interacting protein
|
|
chr1_+_1994943
|
1.215
|
NM_001033581
|
PRKCZ
|
protein kinase C, zeta
|
|
chr3_+_197851052
|
1.211
|
NM_198565
|
LRRC33
|
leucine rich repeat containing 33
|
|
chr6_+_105511615
|
1.200
|
NM_001004317
|
LIN28B
|
lin-28 homolog B (C. elegans)
|
|
chr1_-_152394215
|
1.200
|
NM_001159484
NM_207308
|
NUP210L
|
nucleoporin 210kDa-like
|
|
chrX_-_151996613
|
1.181
|
NM_032882
|
PNMA6A
|
paraneoplastic antigen like 6A
|
|
chr7_-_129379947
|
1.175
|
NM_003344
NM_182697
|
UBE2H
|
ubiquitin-conjugating enzyme E2H (UBC8 homolog, yeast)
|
|
chr7_-_112515068
|
1.163
|
NM_001146265
NM_018970
|
GPR85
|
G protein-coupled receptor 85
|
|
chr19_+_16048315
|
1.159
|
|
TPM4
|
tropomyosin 4
|
|
chr6_-_46401174
|
1.133
|
|
RCAN2
|
regulator of calcineurin 2
|
|
chr6_+_44322764
|
1.130
|
NM_007355
|
HSP90AB1
|
heat shock protein 90kDa alpha (cytosolic), class B member 1
|
|
chr2_-_219882302
|
1.127
|
NM_002846
|
PTPRN
|
protein tyrosine phosphatase, receptor type, N
|
|
chr7_-_112515035
|
1.124
|
|
GPR85
|
G protein-coupled receptor 85
|
|
chr19_-_54231884
|
1.122
|
NM_033377
|
CGB1
CGB2
|
chorionic gonadotropin, beta polypeptide 1
chorionic gonadotropin, beta polypeptide 2
|
|
chr4_-_111777651
|
1.117
|
|
PITX2
|
paired-like homeodomain 2
|
|
chr1_-_149955870
|
1.113
|
NM_007185
|
CELF3
|
CUGBP, Elav-like family member 3
|
|
chr2_-_219882445
|
1.113
|
|
PTPRN
|
protein tyrosine phosphatase, receptor type, N
|
|
chrX_+_148671394
|
1.106
|
NM_005365
NM_001080790
|
MAGEA9
MAGEA9B
|
melanoma antigen family A, 9
melanoma antigen family A, 9B
|
|
chr6_-_32184975
|
1.102
|
NM_019105
|
TNXB
|
tenascin XB
|
|
chr13_+_110771015
|
1.101
|
NM_152324
|
C13orf16
|
chromosome 13 open reading frame 16
|
|
chr6_-_29703725
|
1.097
|
|
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1
|
|
chr7_-_112514990
|
1.094
|
|
GPR85
|
G protein-coupled receptor 85
|
|
chr1_+_46151845
|
1.092
|
|
MAST2
|
microtubule associated serine/threonine kinase 2
|
|
chr1_+_110328850
|
1.085
|
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr19_+_10404110
|
1.083
|
NM_001111309
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr16_-_45339605
|
1.076
|
NM_182493
|
MYLK3
|
myosin light chain kinase 3
|
|
chr11_-_8636958
|
1.076
|
NM_014818
|
TRIM66
|
tripartite motif containing 66
|
|
chr6_+_136214495
|
1.068
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr19_+_60487578
|
1.063
|
|
BRSK1
|
BR serine/threonine kinase 1
|
|
chr6_-_166321393
|
1.062
|
|
C6orf176
|
chromosome 6 open reading frame 176
|
|
chr7_+_128099581
|
1.052
|
NM_001012454
NM_001128926
|
FAM71F2
|
family with sequence similarity 71, member F2
|
|
chr11_-_111287657
|
1.049
|
NM_001885
|
CRYAB
|
crystallin, alpha B
|
|
chr12_+_131194962
|
1.034
|
|
|
|
|
chr4_-_111777584
|
1.028
|
|
PITX2
|
paired-like homeodomain 2
|
|
chr6_-_46401363
|
1.027
|
NM_005822
|
RCAN2
|
regulator of calcineurin 2
|
|
chr2_+_74635019
|
1.015
|
NM_001381
|
DOK1
|
docking protein 1, 62kDa (downstream of tyrosine kinase 1)
|
|
chr7_-_158314945
|
1.014
|
|
ESYT2
|
extended synaptotagmin-like protein 2
|
|
chr7_+_5598979
|
1.002
|
NM_003088
|
FSCN1
|
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus)
|
|
chr2_-_74455161
|
1.002
|
NM_001135041
NM_023019
|
DCTN1
|
dynactin 1
|
|
chr15_-_81037554
|
0.997
|
NM_001079533
NM_001079534
NM_001079535
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1
|
|
chr11_-_47356479
|
0.996
|
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene spi1
|
|
chr11_-_47356650
|
0.993
|
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene spi1
|
|
chr6_+_44322831
|
0.984
|
|
HSP90AB1
|
heat shock protein 90kDa alpha (cytosolic), class B member 1
|
|
chr17_-_13445942
|
0.980
|
NM_006042
|
HS3ST3A1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1
|
|
chr7_-_23537968
|
0.979
|
NM_013293
|
TRA2A
|
transformer 2 alpha homolog (Drosophila)
|
|
chr12_-_131194794
|
0.967
|
NM_175066
|
DDX51
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 51
|
|
chr11_-_47356642
|
0.966
|
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene spi1
|
|
chr11_-_47356672
|
0.966
|
NM_001080547
NM_003120
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene spi1
|
|
chr10_+_18669619
|
0.964
|
NM_201590
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit
|
|
chr7_+_5598964
|
0.962
|
|
FSCN1
|
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus)
|
|
chr12_-_121946635
|
0.950
|
NM_024667
|
VPS37B
|
vacuolar protein sorting 37 homolog B (S. cerevisiae)
|
|
chr11_-_47356562
|
0.949
|
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene spi1
|
|
chr21_-_43671355
|
0.949
|
NM_173354
|
SIK1
|
salt-inducible kinase 1
|
|
chr1_+_110328941
|
0.948
|
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr1_+_1104939
|
0.934
|
NM_153254
|
TTLL10
|
tubulin tyrosine ligase-like family, member 10
|
|
chr19_+_1892184
|
0.928
|
|
CSNK1G2
|
casein kinase 1, gamma 2
|
|
chr12_-_121946581
|
0.923
|
|
VPS37B
|
vacuolar protein sorting 37 homolog B (S. cerevisiae)
|
|
chr1_+_22251706
|
0.921
|
NM_001039802
NM_001791
NM_044472
|
CDC42
|
cell division cycle 42 (GTP binding protein, 25kDa)
|
|
chr1_-_172103745
|
0.918
|
|
GAS5
|
growth arrest-specific 5 (non-protein coding)
|
|
chr8_+_23442331
|
0.917
|
|
SLC25A37
|
solute carrier family 25, member 37
|
|
chr1_-_180822686
|
0.909
|
NM_021133
|
RNASEL
|
ribonuclease L (2',5'-oligoisoadenylate synthetase-dependent)
|
|
chr11_+_18372687
|
0.901
|
|
LDHA
|
lactate dehydrogenase A
|
|
chr21_+_43903803
|
0.900
|
NM_015056
|
RRP1B
|
ribosomal RNA processing 1 homolog B (S. cerevisiae)
|
|
chr22_-_18807805
|
0.899
|
|
PI4KAP2
PI4KAP1
|
phosphatidylinositol 4-kinase, catalytic, alpha pseudogene 2
phosphatidylinositol 4-kinase, catalytic, alpha pseudogene 1
|
|
chr3_+_142587973
|
0.898
|
|
|
|
|
chr11_-_62213577
|
0.893
|
|
LRRN4CL
|
LRRN4 C-terminal like
|
|
chr2_-_26554413
|
0.892
|
NM_004802
NM_194322
NM_194323
|
OTOF
|
otoferlin
|
|
chr19_+_16048047
|
0.886
|
NM_003290
|
TPM4
|
tropomyosin 4
|
|
chr12_+_131194931
|
0.884
|
NM_024078
|
NOC4L
|
nucleolar complex associated 4 homolog (S. cerevisiae)
|
|
chr17_+_22645232
|
0.880
|
NM_015626
NM_134265
|
WSB1
|
WD repeat and SOCS box containing 1
|
|
chr2_-_96899396
|
0.879
|
NM_017789
|
SEMA4C
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C
|
|
chr1_+_196874867
|
0.871
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr8_-_102872354
|
0.871
|
|
NCALD
|
neurocalcin delta
|
|
chr7_+_116447499
|
0.869
|
|
ST7
|
suppression of tumorigenicity 7
|
|
chr19_+_55671594
|
0.868
|
|
C19orf63
|
chromosome 19 open reading frame 63
|
|
chr1_+_9522133
|
0.864
|
|
SLC25A33
|
solute carrier family 25, member 33
|
|
chr19_+_60487345
|
0.857
|
NM_032430
|
BRSK1
|
BR serine/threonine kinase 1
|
|
chr21_+_34112456
|
0.856
|
|
ITSN1
|
intersectin 1 (SH3 domain protein)
|
|
chr1_+_110328830
|
0.852
|
NM_006621
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr18_+_52965290
|
0.852
|
|
BOD1P
|
biorientation of chromosomes in cell division 1 pseudogene
|
|
chr9_-_14076901
|
0.850
|
|
NFIB
|
nuclear factor I/B
|
|
chr4_+_75529716
|
0.843
|
NM_001657
|
AREG
|
amphiregulin
|
|
chr11_+_18372682
|
0.842
|
|
LDHA
|
lactate dehydrogenase A
|
|
chr19_+_1892157
|
0.839
|
NM_001319
|
CSNK1G2
|
casein kinase 1, gamma 2
|
|
chr6_+_39868116
|
0.839
|
|
|
|
|
chr19_+_47593137
|
0.839
|
|
|
|
|
chr1_+_196874839
|
0.827
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr7_-_137271191
|
0.826
|
|
CREB3L2
|
cAMP responsive element binding protein 3-like 2
|
|
chr2_+_151922350
|
0.824
|
NM_007115
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6
|
|
chr1_-_152431175
|
0.823
|
NM_152263
|
TPM3
|
tropomyosin 3
|
|
chr11_+_18372689
|
0.823
|
|
LDHA
|
lactate dehydrogenase A
|
|
chr19_+_55671584
|
0.812
|
|
C19orf63
|
chromosome 19 open reading frame 63
|
|
chr6_+_45404031
|
0.810
|
NM_001015051
NM_001024630
|
RUNX2
|
runt-related transcription factor 2
|
|
chr17_-_44058612
|
0.809
|
NM_024017
|
HOXB9
|
homeobox B9
|
|
chr7_+_100302885
|
0.806
|
NM_003302
|
TRIP6
|
thyroid hormone receptor interactor 6
|
|
chr6_-_15356865
|
0.804
|
|
|
|
|
chr11_+_18372708
|
0.798
|
|
LDHA
|
lactate dehydrogenase A
|
|
chr22_-_18484428
|
0.795
|
|
TRMT2A
|
TRM2 tRNA methyltransferase 2 homolog A (S. cerevisiae)
|
|
chr10_-_50012058
|
0.794
|
NM_001164484
|
FAM170B
|
family with sequence similarity 170, member B
|
|
chr1_-_51660328
|
0.792
|
NM_001159969
|
EPS15
|
epidermal growth factor receptor pathway substrate 15
|
|
chr21_-_43903755
|
0.785
|
NM_007031
|
HSF2BP
|
heat shock transcription factor 2 binding protein
|
|
chrX_+_151991474
|
0.783
|
NM_001170944
NM_032882
|
LOC100287428
PNMA6A
|
paraneoplastic antigen-like protein 6A related gene
paraneoplastic antigen like 6A
|
|
chr1_-_240228987
|
0.782
|
NM_001004343
|
MAP1LC3C
|
microtubule-associated protein 1 light chain 3 gamma
|
|
chr19_+_55671600
|
0.779
|
|
C19orf63
|
chromosome 19 open reading frame 63
|
|
chr3_-_33734544
|
0.777
|
|
CLASP2
|
cytoplasmic linker associated protein 2
|
|
chr20_+_55569542
|
0.774
|
NM_002591
|
PCK1
|
phosphoenolpyruvate carboxykinase 1 (soluble)
|
|
chr7_-_100595552
|
0.774
|
NM_003378
|
VGF
|
VGF nerve growth factor inducible
|
|
chr9_+_131291224
|
0.773
|
|
LOC100506190
|
hypothetical LOC100506190
|
|
chr1_+_110328955
|
0.771
|
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chrX_+_9391314
|
0.770
|
NM_001139466
NM_001139467
NM_001139468
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr8_-_102872372
|
0.761
|
|
NCALD
|
neurocalcin delta
|
|
chr1_+_226394589
|
0.760
|
|
GUK1
|
guanylate kinase 1
|
|
chr7_-_25131427
|
0.755
|
NM_018947
|
CYCS
|
cytochrome c, somatic
|
|
chr6_+_39868126
|
0.751
|
|
DAAM2
|
dishevelled associated activator of morphogenesis 2
|
|
chr7_+_65596078
|
0.747
|
|
|
|
|
chr7_+_30927439
|
0.743
|
NM_001185060
NM_001185061
NM_001185062
|
AQP1
|
aquaporin 1 (Colton blood group)
|
|
chr19_+_54067450
|
0.741
|
NM_014330
|
PPP1R15A
|
protein phosphatase 1, regulatory (inhibitor) subunit 15A
|
|
chr13_-_27967215
|
0.736
|
NM_001159920
NM_001160030
NM_001160031
NM_002019
|
FLT1
|
fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor)
|
|
chr19_+_54067489
|
0.733
|
|
PPP1R15A
|
protein phosphatase 1, regulatory (inhibitor) subunit 15A
|
|
chr2_-_156897286
|
0.730
|
NM_006186
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2
|
|
chr12_+_52665196
|
0.730
|
NM_017409
|
HOXC10
|
homeobox C10
|
|
chr19_-_63758270
|
0.727
|
|
CHMP2A
|
chromatin modifying protein 2A
|
|
chr3_+_148593911
|
0.726
|
|
|
|
|
chr8_-_145723209
|
0.722
|
NM_001024678
|
LRRC24
|
leucine rich repeat containing 24
|
|
chr1_+_196874759
|
0.722
|
NM_002838
NM_080921
NM_080923
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chrX_+_153325643
|
0.722
|
NM_004699
|
FAM50A
|
family with sequence similarity 50, member A
|
|
chr1_+_9522107
|
0.719
|
NM_032315
|
SLC25A33
|
solute carrier family 25, member 33
|
|
chr1_+_226394626
|
0.717
|
|
GUK1
|
guanylate kinase 1
|
|
chr20_+_54533191
|
0.716
|
NM_001012971
|
C20orf106
C20orf107
|
chromosome 20 open reading frame 106
chromosome 20 open reading frame 107
|
|
chr8_-_21827057
|
0.711
|
NM_003974
|
DOK2
|
docking protein 2, 56kDa
|
|
chr7_-_37922902
|
0.709
|
|
SFRP4
|
secreted frizzled-related protein 4
|
|
chr10_-_7491204
|
0.707
|
NM_001018039
|
SFMBT2
|
Scm-like with four mbt domains 2
|
|
chr6_-_31779068
|
0.707
|
|
ABHD16A
|
abhydrolase domain containing 16A
|
|
chr8_-_18710575
|
0.706
|
NM_206909
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr6_+_15357013
|
0.706
|
|
JARID2
|
jumonji, AT rich interactive domain 2
|
|
chrX_+_153325684
|
0.706
|
|
FAM50A
|
family with sequence similarity 50, member A
|
|
chr5_+_118993152
|
0.706
|
NM_001163991
NM_182761
|
FAM170A
|
family with sequence similarity 170, member A
|
|
chr11_+_117362272
|
0.704
|
NM_001558
|
IL10RA
|
interleukin 10 receptor, alpha
|
|
chr7_-_37923040
|
0.704
|
NM_003014
|
SFRP4
|
secreted frizzled-related protein 4
|
|
chr4_+_75699652
|
0.703
|
NM_001657
|
AREG
|
amphiregulin
|
|
chr8_-_102033409
|
0.700
|
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide
|
|
chr1_+_201541241
|
0.698
|
|
BTG2
|
BTG family, member 2
|
|
chr18_+_22060383
|
0.698
|
NM_005640
|
TAF4B
|
TAF4b RNA polymerase II, TATA box binding protein (TBP)-associated factor, 105kDa
|
|
chr9_+_139152595
|
0.697
|
|
GRIN1
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1
|
|
chr19_-_63758184
|
0.697
|
|
CHMP2A
|
chromatin modifying protein 2A
|
|
chr11_-_62213946
|
0.696
|
NM_203422
|
LRRN4CL
|
LRRN4 C-terminal like
|
|
chr22_+_23333610
|
0.696
|
NM_001032365
|
GGT1
|
gamma-glutamyltransferase 1
|
|
chr19_+_54996613
|
0.694
|
|
|
|
|
chr1_+_111217244
|
0.692
|
NM_000560
|
CD53
|
CD53 molecule
|
|
chr8_-_102033444
|
0.691
|
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide
|
|
chrX_+_154880439
|
0.687
|
NM_002186
|
IL9R
|
interleukin 9 receptor
|
|
chr8_-_102033372
|
0.686
|
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide
|
|
chr21_-_38210531
|
0.685
|
NM_002240
|
KCNJ6
|
potassium inwardly-rectifying channel, subfamily J, member 6
|
|
chr8_-_102033434
|
0.684
|
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide
|
|
chr20_-_55718351
|
0.683
|
NM_020182
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1
|
|
chr19_-_55671814
|
0.683
|
NM_138411
|
FAM71E1
|
family with sequence similarity 71, member E1
|
|
chr1_+_226394605
|
0.683
|
|
GUK1
|
guanylate kinase 1
|
|
chr6_-_31779082
|
0.682
|
|
ABHD16A
|
abhydrolase domain containing 16A
|
|
chr19_-_6432765
|
0.680
|
NM_024898
|
DENND1C
|
DENN/MADD domain containing 1C
|
|
chr20_+_17155617
|
0.677
|
NM_002594
|
PCSK2
|
proprotein convertase subtilisin/kexin type 2
|
|
chr3_-_187138470
|
0.677
|
|
TRA2B
|
transformer 2 beta homolog (Drosophila)
|