|
chr2_+_11597530
|
11.678
|
NM_148903
|
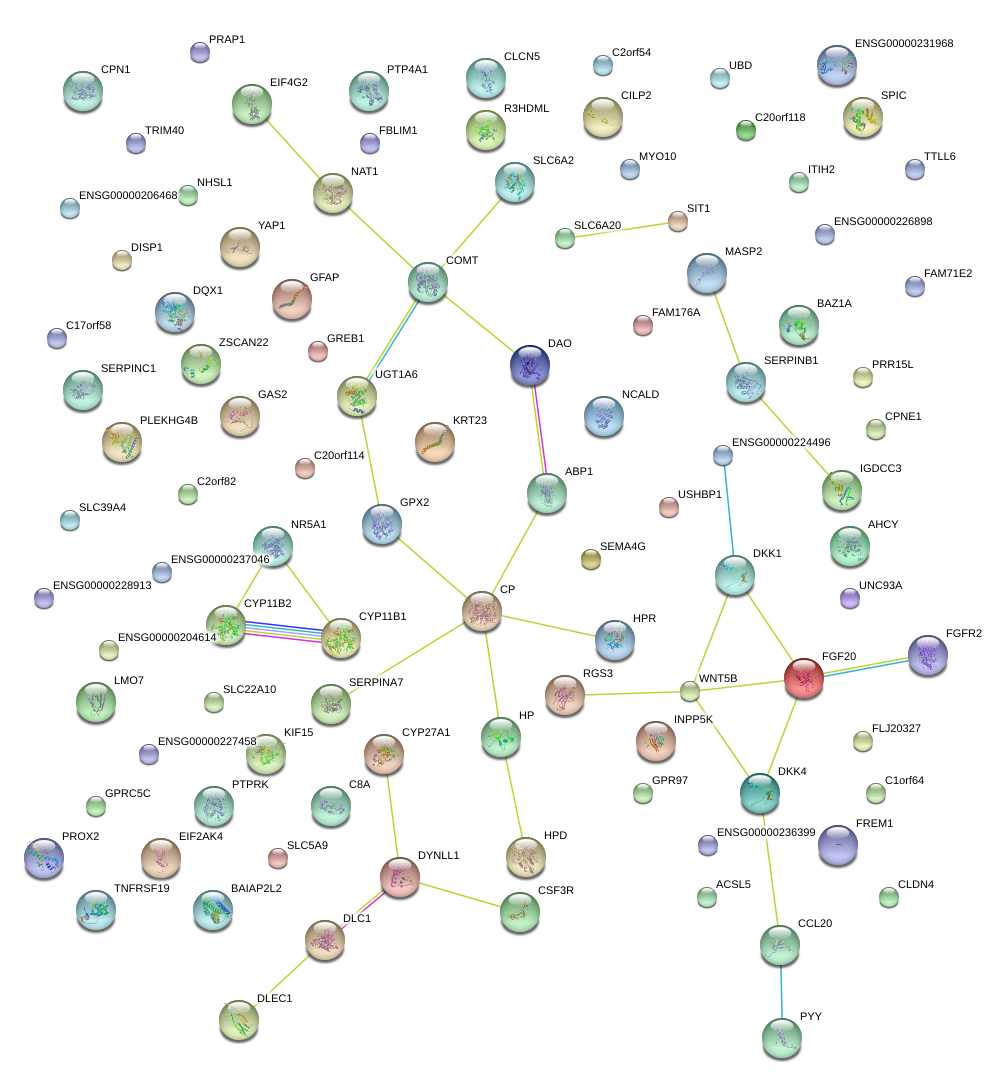
GREB1
|
growth regulation by estrogen in breast cancer 1
|
|
chr6_+_167624792
|
5.318
|
NM_001143947
NM_018974
|
UNC93A
|
unc-93 homolog A (C. elegans)
|
|
chr7_+_72883128
|
4.982
|
NM_001305
|
CLDN4
|
claudin 4
|
|
chr2_+_233443237
|
4.955
|
NM_206895
|
C2orf82
|
chromosome 2 open reading frame 82
|
|
chr12_-_120781002
|
4.796
|
NM_002150
|
HPD
|
4-hydroxyphenylpyruvate dioxygenase
|
|
chr5_-_16791346
|
4.437
|
|
MYO10
|
myosin X
|
|
chr3_-_150422474
|
4.172
|
NM_000096
|
CP
|
ceruloplasmin (ferroxidase)
|
|
chr17_-_76985538
|
4.095
|
|
|
|
|
chr2_+_228386800
|
4.058
|
NM_001130046
NM_004591
|
CCL20
|
chemokine (C-C motif) ligand 20
|
|
chr20_+_34937928
|
3.973
|
NM_080628
|
C20orf118
|
chromosome 20 open reading frame 118
|
|
chr10_+_7785347
|
3.556
|
|
ITIH2
|
inter-alpha (globulin) inhibitor H2
|
|
chr17_-_36347197
|
3.549
|
NM_015515
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr20_-_42457717
|
3.509
|
|
|
|
|
chr17_-_43390108
|
3.452
|
NM_024320
|
PRR15L
|
proline rich 15-like
|
|
chr10_+_7785360
|
3.411
|
|
ITIH2
|
inter-alpha (globulin) inhibitor H2
|
|
chr14_-_64479198
|
3.358
|
NM_002083
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal)
|
|
chr13_+_75232797
|
3.325
|
NM_015842
|
LMO7
|
LIM domain 7
|
|
chr16_+_56259657
|
3.273
|
NM_170776
|
GPR97
|
G protein-coupled receptor 97
|
|
chr2_+_11600238
|
3.261
|
|
GREB1
|
growth regulation by estrogen in breast cancer 1
|
|
chr15_+_38082041
|
3.214
|
|
EIF2AK4
|
eukaryotic translation initiation factor 2 alpha kinase 4
|
|
chr6_-_138935359
|
3.154
|
NM_020464
|
NHSL1
|
NHS-like 1
|
|
chr1_+_16203317
|
3.140
|
NM_178840
|
C1orf64
|
chromosome 1 open reading frame 64
|
|
chr20_-_32363268
|
3.056
|
NM_001161766
|
AHCY
|
adenosylhomocysteinase
|
|
chr9_+_115265827
|
3.040
|
NM_017790
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr2_+_11600287
|
3.020
|
NM_033090
|
GREB1
|
growth regulation by estrogen in breast cancer 1
|
|
chr10_+_7785331
|
2.989
|
|
ITIH2
|
inter-alpha (globulin) inhibitor H2
|
|
chr6_+_131190237
|
2.962
|
NM_001195597
|
LOC285733
|
hypothetical LOC285733
|
|
chr8_-_145613034
|
2.957
|
|
SLC39A4
|
solute carrier family 39 (zinc transporter), member 4
|
|
chr2_-_241484245
|
2.847
|
NM_001085437
|
C2orf54
|
chromosome 2 open reading frame 54
|
|
chr20_+_31334601
|
2.832
|
NM_033197
|
C20orf114
|
chromosome 20 open reading frame 114
|
|
chr20_+_42399039
|
2.729
|
NM_178491
|
R3HDML
|
R3H domain containing-like
|
|
chr11_+_22646229
|
2.679
|
NM_177553
|
GAS2
|
growth arrest-specific 2
|
|
chr10_+_53744043
|
2.660
|
NM_012242
|
DKK1
|
dickkopf homolog 1 (Xenopus laevis)
|
|
chr2_-_74606819
|
2.641
|
NM_133637
|
DQX1
|
DEAQ box RNA-dependent ATPase 1
|
|
chr10_+_7785235
|
2.618
|
NM_002216
|
ITIH2
|
inter-alpha (globulin) inhibitor H2
|
|
chr11_+_62813947
|
2.611
|
NM_001039752
|
SLC22A10
|
solute carrier family 22, member 10
|
|
chr12_+_1608672
|
2.606
|
NM_032642
|
WNT5B
|
wingless-type MMTV integration site family, member 5B
|
|
chr10_+_7785290
|
2.501
|
|
ITIH2
|
inter-alpha (globulin) inhibitor H2
|
|
chr13_+_23042722
|
2.497
|
NM_148957
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19
|
|
chr10_+_135010833
|
2.463
|
NM_001145201
NM_145202
|
PRAP1
|
proline-rich acidic protein 1
|
|
chr10_+_114144512
|
2.443
|
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chrX_-_105169370
|
2.393
|
NM_000354
|
SERPINA7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7
|
|
chr16_+_70646008
|
2.392
|
NM_001126102
NM_005143
|
HP
|
haptoglobin
|
|
chr1_+_48460943
|
2.386
|
NM_001011547
NM_001135181
|
SLC5A9
|
solute carrier family 5 (sodium/glucose cotransporter), member 9
|
|
chr17_-_40348378
|
2.357
|
NM_001131019
NM_002055
|
GFAP
|
glial fibrillary acidic protein
|
|
chr10_+_102722275
|
2.336
|
NM_017893
|
SEMA4G
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G
|
|
chr3_+_152970904
|
2.296
|
|
LOC201651
|
arylacetamide deacetylase (esterase) pseudogene
|
|
chr6_-_2785682
|
2.261
|
|
SERPINB1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1
|
|
chr1_+_15957832
|
2.241
|
NM_017556
|
FBLIM1
|
filamin binding LIM protein 1
|
|
chr8_-_42353719
|
2.238
|
NM_014420
|
DKK4
|
dickkopf homolog 4 (Xenopus laevis)
|
|
chr11_+_101488380
|
2.200
|
NM_001195045
|
YAP1
|
Yes-associated protein 1
|
|
chr17_-_36346240
|
2.200
|
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr2_+_219354948
|
2.190
|
|
CYP27A1
|
cytochrome P450, family 27, subfamily A, polypeptide 1
|
|
chrX_+_49718954
|
2.166
|
NM_000084
|
CLCN5
|
chloride channel 5
|
|
chr17_+_7900926
|
2.159
|
|
|
|
|
chr10_-_123346148
|
2.143
|
NM_001144915
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr17_+_69939806
|
2.130
|
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C
|
|
chr2_-_75641535
|
2.106
|
|
FAM176A
|
family with sequence similarity 176, member A
|
|
chr17_-_44226595
|
2.105
|
NM_173623
|
TTLL6
|
tubulin tyrosine ligase-like family, member 6
|
|
chr2_-_75641587
|
2.085
|
NM_001135032
|
FAM176A
|
family with sequence similarity 176, member A
|
|
chr9_-_126309510
|
2.064
|
NM_004959
|
NR5A1
|
nuclear receptor subfamily 5, group A, member 1
|
|
chr9_-_35640852
|
2.064
|
NM_014450
|
SIT1
|
signaling threshold regulating transmembrane adaptor 1
|
|
chr1_+_57093030
|
2.063
|
NM_000562
|
C8A
|
complement component 8, alpha polypeptide
|
|
chr22_+_18330049
|
2.058
|
NM_007310
|
COMT
|
catechol-O-methyltransferase
|
|
chr9_-_14858974
|
2.031
|
|
FREM1
|
FRAS1 related extracellular matrix 1
|
|
chr17_-_1359745
|
2.015
|
|
INPP5K
|
inositol polyphosphate-5-phosphatase K
|
|
chr15_-_63457378
|
1.932
|
NM_004884
|
IGDCC3
|
immunoglobulin superfamily, DCC subclass, member 3
|
|
chr6_-_29635456
|
1.871
|
|
UBD
|
ubiquitin D
|
|
chr19_-_17236522
|
1.836
|
NM_031941
|
USHBP1
|
Usher syndrome 1C binding protein 1
|
|
chr6_-_29635680
|
1.835
|
NM_006398
|
UBD
|
ubiquitin D
|
|
chr8_+_18072250
|
1.831
|
NM_001160179
|
NAT1
|
N-acetyltransferase 1 (arylamine N-acetyltransferase)
|
|
chr8_+_121892719
|
1.829
|
|
|
|
|
chr1_-_36721501
|
1.826
|
NM_000760
NM_156039
NM_172313
|
CSF3R
|
colony stimulating factor 3 receptor (granulocyte)
|
|
chr6_-_128883125
|
1.813
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chr8_-_16903911
|
1.811
|
NM_019851
|
FGF20
|
fibroblast growth factor 20
|
|
chr10_-_101831579
|
1.805
|
NM_001308
|
CPN1
|
carboxypeptidase N, polypeptide 1
|
|
chr1_-_172153087
|
1.793
|
NM_000488
|
SERPINC1
|
serpin peptidase inhibitor, clade C (antithrombin), member 1
|
|
chr14_-_34413831
|
1.789
|
|
BAZ1A
|
bromodomain adjacent to zinc finger domain, 1A
|
|
chr12_+_107797985
|
1.777
|
NM_001917
|
DAO
|
D-amino-acid oxidase
|
|
chr1_+_221168388
|
1.764
|
NM_032890
|
DISP1
|
dispatched homolog 1 (Drosophila)
|
|
chr6_+_30212488
|
1.747
|
NM_138700
|
TRIM40
|
tripartite motif containing 40
|
|
chr6_+_64339869
|
1.745
|
NM_003463
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1
|
|
chr1_-_11029737
|
1.734
|
NM_006610
NM_139208
|
MASP2
|
mannan-binding lectin serine peptidase 2
|
|
chr17_-_39437362
|
1.672
|
NM_004160
|
PYY
|
peptide YY
|
|
chr11_+_46697312
|
1.659
|
NM_000506
|
F2
|
coagulation factor II (thrombin)
|
|
chr13_+_109936080
|
1.659
|
|
COL4A2
|
collagen, type IV, alpha 2
|
|
chr17_+_72795567
|
1.654
|
NM_001113492
|
SEPT9
|
septin 9
|
|
chr17_-_23721468
|
1.642
|
|
VTN
|
vitronectin
|
|
chr14_-_34414277
|
1.628
|
|
BAZ1A
|
bromodomain adjacent to zinc finger domain, 1A
|
|
chr16_-_71639670
|
1.626
|
NM_006885
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr2_+_97696462
|
1.623
|
NM_001079
|
ZAP70
|
zeta-chain (TCR) associated protein kinase 70kDa
|
|
chr14_-_30746577
|
1.623
|
|
|
|
|
chr6_+_35812836
|
1.616
|
NM_145028
|
C6orf81
|
chromosome 6 open reading frame 81
|
|
chr20_-_17459973
|
1.602
|
NM_001195
|
BFSP1
|
beaded filament structural protein 1, filensin
|
|
chr15_-_87565813
|
1.580
|
NM_000326
|
RLBP1
|
retinaldehyde binding protein 1
|
|
chr19_-_50506883
|
1.568
|
|
CKM
|
creatine kinase, muscle
|
|
chr21_-_42608531
|
1.565
|
|
TFF3
|
trefoil factor 3 (intestinal)
|
|
chr7_+_29200636
|
1.563
|
NM_004067
|
CHN2
|
chimerin (chimaerin) 2
|
|
chr10_-_73518284
|
1.560
|
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2
|
|
chr7_+_142692165
|
1.553
|
NM_153345
|
TMEM139
|
transmembrane protein 139
|
|
chr11_+_46697345
|
1.527
|
|
F2
|
coagulation factor II (thrombin)
|
|
chr1_+_2026014
|
1.525
|
NM_001033582
|
PRKCZ
|
protein kinase C, zeta
|
|
chr7_+_94953148
|
1.516
|
NM_016116
NM_145872
|
ASB4
|
ankyrin repeat and SOCS box containing 4
|
|
chr1_-_172153024
|
1.508
|
|
SERPINC1
|
serpin peptidase inhibitor, clade C (antithrombin), member 1
|
|
chr7_+_100169184
|
1.506
|
NM_003386
NM_173059
|
ZAN
|
zonadhesin
|
|
chr14_-_30746431
|
1.501
|
NM_015382
|
HECTD1
|
HECT domain containing 1
|
|
chr14_-_34414201
|
1.488
|
|
BAZ1A
|
bromodomain adjacent to zinc finger domain, 1A
|
|
chr11_+_46697335
|
1.484
|
|
F2
|
coagulation factor II (thrombin)
|
|
chr6_+_167456230
|
1.474
|
NM_031409
|
CCR6
|
chemokine (C-C motif) receptor 6
|
|
chr2_+_69093641
|
1.473
|
NM_018153
NM_032208
NM_053034
|
ANTXR1
|
anthrax toxin receptor 1
|
|
chr18_-_24010944
|
1.467
|
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal)
|
|
chr17_+_7474177
|
1.459
|
NM_001040
NM_001146279
NM_001146280
NM_001146281
|
SHBG
|
sex hormone-binding globulin
|
|
chr1_-_108032645
|
1.456
|
NM_001079874
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr14_-_34414241
|
1.455
|
|
BAZ1A
|
bromodomain adjacent to zinc finger domain, 1A
|
|
chr17_+_35425139
|
1.440
|
NM_000759
NM_001178147
NM_172219
NM_172220
|
CSF3
|
colony stimulating factor 3 (granulocyte)
|
|
chr20_+_39199263
|
1.437
|
|
PLCG1
|
phospholipase C, gamma 1
|
|
chr4_+_17125885
|
1.419
|
NM_001079827
|
CLRN2
|
clarin 2
|
|
chr14_-_22521544
|
1.410
|
NM_032876
|
JUB
|
jub, ajuba homolog (Xenopus laevis)
|
|
chr11_-_124311106
|
1.401
|
|
HEPACAM
|
hepatic and glial cell adhesion molecule
|
|
chr1_-_110108048
|
1.395
|
NM_024526
NM_133181
NM_139053
|
EPS8L3
|
EPS8-like 3
|
|
chr7_+_139175420
|
1.392
|
NM_001061
NM_001166253
NM_030984
|
TBXAS1
|
thromboxane A synthase 1 (platelet)
|
|
chr11_+_74548472
|
1.381
|
NM_001145211
|
SLCO2B1
|
solute carrier organic anion transporter family, member 2B1
|
|
chr13_+_75108450
|
1.377
|
|
LMO7
|
LIM domain 7
|
|
chr1_-_1346465
|
1.366
|
NM_001145210
|
LOC441869
|
hypothetical protein LOC441869
|
|
chr10_-_52315390
|
1.364
|
|
A1CF
|
APOBEC1 complementation factor
|
|
chr8_-_121893196
|
1.354
|
|
|
|
|
chr2_+_135312655
|
1.352
|
NM_138326
|
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase
|
|
chr8_-_121892877
|
1.333
|
|
SNTB1
|
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1)
|
|
chr11_+_62893836
|
1.333
|
NM_080866
|
SLC22A9
|
solute carrier family 22 (organic anion transporter), member 9
|
|
chr9_+_111927601
|
1.329
|
NM_001136562
|
AKAP2
|
A kinase (PRKA) anchor protein 2
|
|
chr2_-_169927258
|
1.326
|
NM_004525
|
LRP2
|
low density lipoprotein receptor-related protein 2
|
|
chr11_-_75599433
|
1.326
|
|
WNT11
|
wingless-type MMTV integration site family, member 11
|
|
chr4_-_158111504
|
1.314
|
|
PDGFC
|
platelet derived growth factor C
|
|
chrX_+_152413646
|
1.311
|
|
BGN
|
biglycan
|
|
chr5_-_58370693
|
1.310
|
NM_001197221
NM_001197222
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr15_+_60146467
|
1.310
|
NM_207322
|
C2CD4A
|
C2 calcium-dependent domain containing 4A
|
|
chr14_-_20642629
|
1.305
|
NM_001102454
|
ZNF219
|
zinc finger protein 219
|
|
chr2_+_158666627
|
1.292
|
NM_173355
|
UPP2
|
uridine phosphorylase 2
|
|
chr9_-_116190038
|
1.273
|
|
AKNA
|
AT-hook transcription factor
|
|
chr20_-_1396336
|
1.261
|
NM_016143
NM_018839
NM_182483
|
NSFL1C
|
NSFL1 (p97) cofactor (p47)
|
|
chr3_+_187813405
|
1.249
|
|
AHSG
|
alpha-2-HS-glycoprotein
|
|
chrX_+_152413540
|
1.244
|
NM_001711
|
BGN
|
biglycan
|
|
chr7_-_27186400
|
1.233
|
NM_153715
|
HOXA10
|
homeobox A10
|
|
chr19_-_18578467
|
1.230
|
|
CRLF1
|
cytokine receptor-like factor 1
|
|
chr19_+_59061422
|
1.230
|
NM_001020820
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr3_-_140677990
|
1.224
|
NM_004164
|
RBP2
|
retinol binding protein 2, cellular
|
|
chr3_-_115960807
|
1.224
|
NM_001164344
NM_001164345
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr2_+_171280046
|
1.215
|
NM_001003845
|
SP5
|
Sp5 transcription factor
|
|
chr19_+_8180179
|
1.202
|
NM_024552
|
LASS4
|
LAG1 homolog, ceramide synthase 4
|
|
chr14_-_34414603
|
1.197
|
NM_013448
NM_182648
|
BAZ1A
|
bromodomain adjacent to zinc finger domain, 1A
|
|
chr19_-_6671585
|
1.189
|
NM_000064
|
C3
|
complement component 3
|
|
chrX_+_46822751
|
1.189
|
|
RGN
|
regucalcin (senescence marker protein-30)
|
|
chr13_-_22905822
|
1.184
|
NM_014363
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin)
|
|
chr10_-_98935369
|
1.183
|
|
SLIT1
|
slit homolog 1 (Drosophila)
|
|
chr6_-_31789824
|
1.178
|
|
|
|
|
chr14_+_38720085
|
1.155
|
|
PNN
|
pinin, desmosome associated protein
|
|
chr17_-_53849888
|
1.138
|
|
RNF43
|
ring finger protein 43
|
|
chr4_-_40911212
|
1.130
|
NM_001166050
NM_004307
NM_173075
|
APBB2
|
amyloid beta (A4) precursor protein-binding, family B, member 2
|
|
chr18_-_24011349
|
1.129
|
NM_001792
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal)
|
|
chr1_-_75854281
|
1.127
|
|
SLC44A5
|
solute carrier family 44, member 5
|
|
chr4_-_7991992
|
1.124
|
NM_001134647
NM_198595
|
AFAP1
|
actin filament associated protein 1
|
|
chr8_+_17478214
|
1.119
|
NM_006207
|
PDGFRL
|
platelet-derived growth factor receptor-like
|
|
chrX_-_55037235
|
1.118
|
NM_002625
|
PFKFB1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1
|
|
chr2_+_70981223
|
1.101
|
NM_012476
|
VAX2
|
ventral anterior homeobox 2
|
|
chr6_+_44076281
|
1.097
|
NM_001171992
NM_153246
|
C6orf223
|
chromosome 6 open reading frame 223
|
|
chr2_+_218992053
|
1.083
|
NM_007127
|
VIL1
|
villin 1
|
|
chr14_-_91111135
|
1.082
|
NM_001080113
|
C14orf184
|
chromosome 14 open reading frame 184
|
|
chr2_+_54537842
|
1.075
|
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1
|
|
chr4_-_158111916
|
1.061
|
|
PDGFC
|
platelet derived growth factor C
|
|
chr17_-_60988226
|
1.054
|
|
AXIN2
|
axin 2
|
|
chr11_-_62753574
|
1.051
|
NM_199352
|
SLC22A25
|
solute carrier family 22, member 25
|
|
chr14_-_93924663
|
1.051
|
|
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1
|
|
chr3_-_121883647
|
1.050
|
|
HGD
|
homogentisate 1,2-dioxygenase
|
|
chr1_+_42701460
|
1.035
|
|
CCDC30
|
coiled-coil domain containing 30
|
|
chr10_-_116434364
|
1.032
|
NM_001003407
NM_001003408
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr11_+_117982422
|
1.031
|
NM_015157
|
PHLDB1
|
pleckstrin homology-like domain, family B, member 1
|
|
chr1_-_153537362
|
1.015
|
NM_181871
|
PKLR
|
pyruvate kinase, liver and RBC
|
|
chr6_-_26038817
|
1.015
|
NM_005835
|
SLC17A2
|
solute carrier family 17 (sodium phosphate), member 2
|
|
chr11_-_34491878
|
1.006
|
NM_001422
|
ELF5
|
E74-like factor 5 (ets domain transcription factor)
|
|
chr5_+_140509803
|
1.003
|
NM_018939
|
PCDHB6
|
protocadherin beta 6
|
|
chr3_-_150858274
|
1.002
|
NM_001168280
NM_015472
|
WWTR1
|
WW domain containing transcription regulator 1
|
|
chr7_-_133794404
|
1.001
|
NM_001628
|
AKR1B1
|
aldo-keto reductase family 1, member B1 (aldose reductase)
|
|
chr19_-_18578631
|
0.990
|
NM_004750
|
CRLF1
|
cytokine receptor-like factor 1
|
|
chr3_-_114643008
|
0.990
|
NM_001164496
NM_018338
|
WDR52
|
WD repeat domain 52
|
|
chr1_-_16217871
|
0.989
|
NM_014424
|
HSPB7
|
heat shock 27kDa protein family, member 7 (cardiovascular)
|
|
chr2_-_135498716
|
0.986
|
NM_001018046
NM_025052
|
YSK4
|
YSK4 Sps1/Ste20-related kinase homolog (S. cerevisiae)
|
|
chr17_-_36376663
|
0.967
|
NM_213656
|
KRT39
|
keratin 39
|
|
chr10_+_104619199
|
0.964
|
NM_020682
|
AS3MT
|
arsenic (+3 oxidation state) methyltransferase
|
|
chr19_-_18578577
|
0.961
|
|
CRLF1
|
cytokine receptor-like factor 1
|
|
chr2_+_233961538
|
0.958
|
NM_003648
|
DGKD
|
diacylglycerol kinase, delta 130kDa
|
|
chr8_+_103889733
|
0.958
|
|
FLJ45248
|
FLJ45248 protein
|
|
chr2_+_219432789
|
0.952
|
NM_006522
|
WNT6
|
wingless-type MMTV integration site family, member 6
|
|
chr7_+_128615948
|
0.952
|
NM_005631
|
SMO
|
smoothened homolog (Drosophila)
|
|
chr17_+_19377732
|
0.946
|
NM_018242
|
SLC47A1
|
solute carrier family 47, member 1
|
|
chr17_+_4621924
|
0.942
|
NM_003963
|
TM4SF5
|
transmembrane 4 L six family member 5
|
|
chr19_-_5789740
|
0.940
|
|
FUT6
|
fucosyltransferase 6 (alpha (1,3) fucosyltransferase)
|
|
chr4_-_158111995
|
0.939
|
NM_016205
|
PDGFC
|
platelet derived growth factor C
|
|
chr11_+_75106512
|
0.930
|
NM_025098
|
MOGAT2
|
monoacylglycerol O-acyltransferase 2
|
|
chrX_+_46822688
|
0.929
|
NM_004683
NM_152869
|
RGN
|
regucalcin (senescence marker protein-30)
|
|
chr12_+_48784057
|
0.925
|
NM_005276
|
GPD1
|
glycerol-3-phosphate dehydrogenase 1 (soluble)
|