|
chr1_-_153098811
|
10.201
|
NM_170782
|
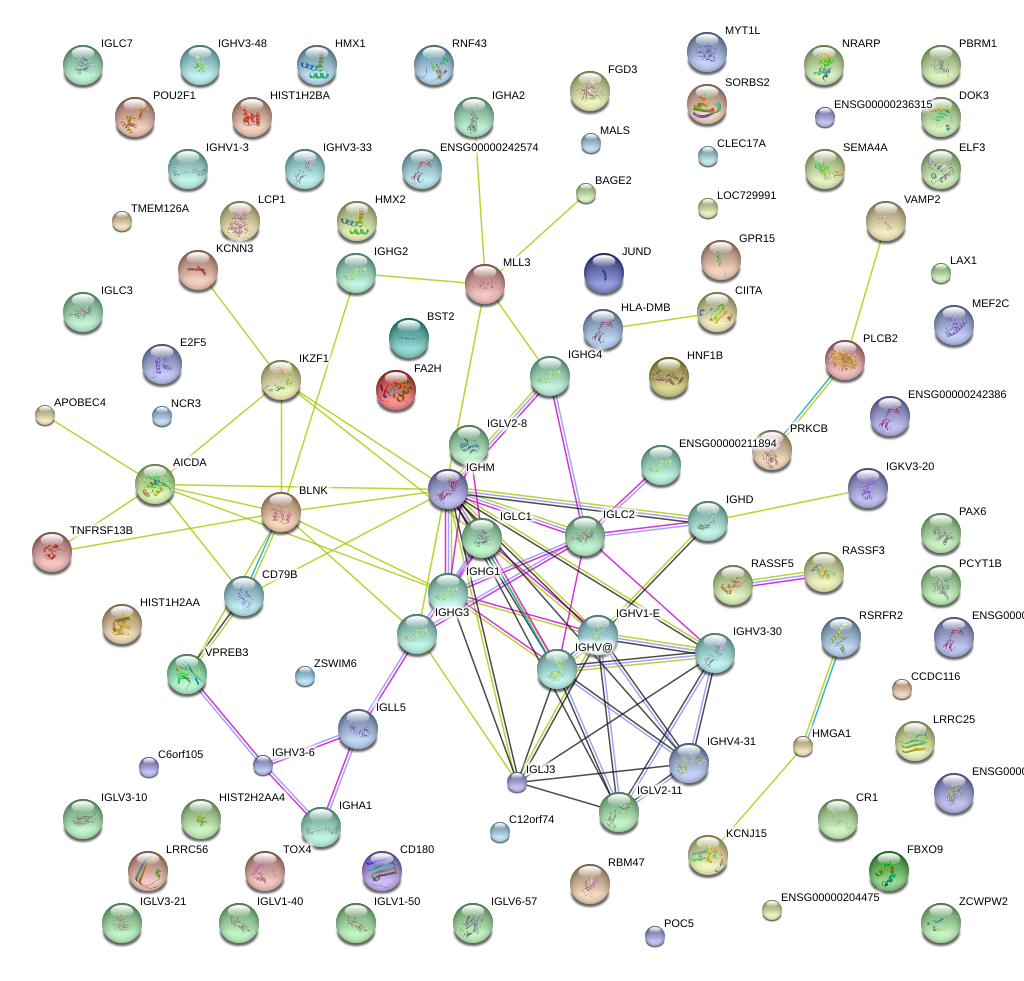
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3
|
|
chr7_+_50318801
|
9.707
|
|
IKZF1
|
IKAROS family zinc finger 1 (Ikaros)
|
|
chr1_+_165565006
|
8.511
|
|
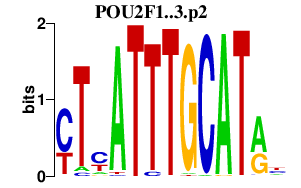
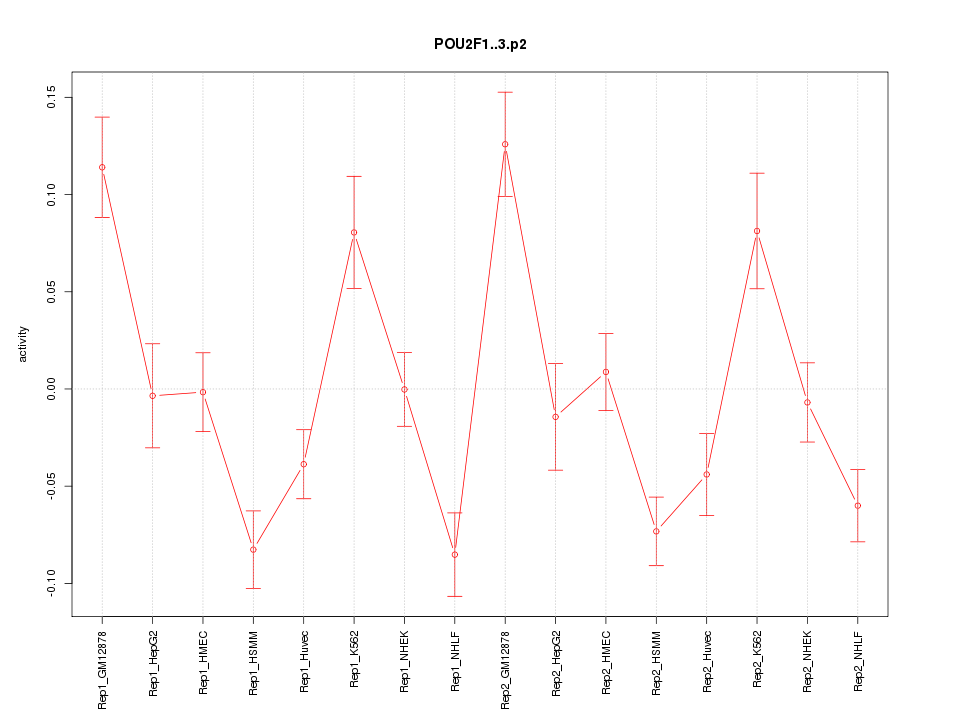
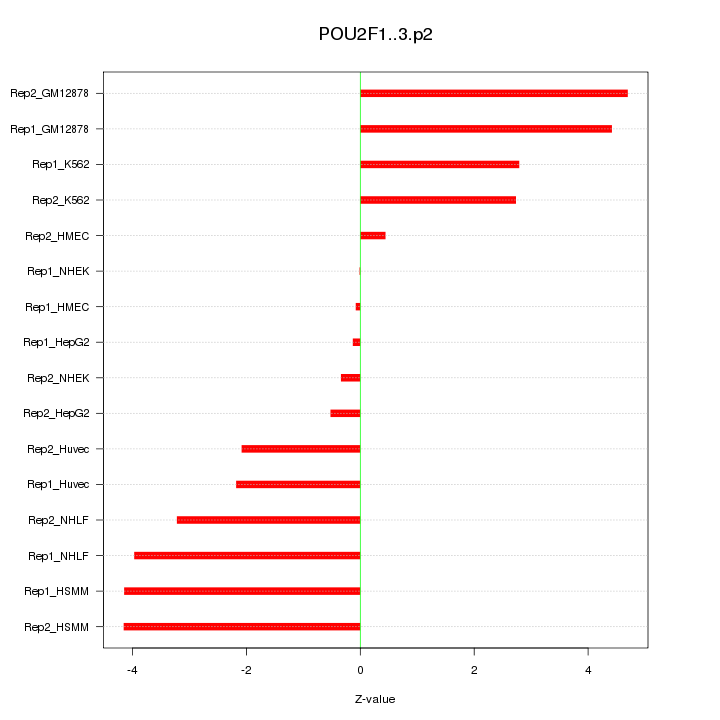
POU2F1
|
POU class 2 homeobox 1
|
|
chr1_+_165564904
|
8.253
|
NM_001198783
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr1_+_154386358
|
6.437
|
NM_001193301
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A
|
|
chr16_+_23754997
|
5.916
|
|
PRKCB
|
protein kinase C, beta
|
|
chr3_+_99733432
|
5.751
|
NM_005290
|
GPR15
|
G protein-coupled receptor 15
|
|
chr22_+_20715340
|
5.532
|
|
|
|
|
chr16_+_23754860
|
5.514
|
|
PRKCB
|
protein kinase C, beta
|
|
chr1_+_202000906
|
5.039
|
NM_001136190
NM_017773
|
LAX1
|
lymphocyte transmembrane adaptor 1
|
|
chr16_+_23754822
|
4.973
|
|
PRKCB
|
protein kinase C, beta
|
|
chr4_-_186970366
|
4.958
|
NM_003603
NM_001145670
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr5_-_88155360
|
4.922
|
NM_001193348
NM_001193349
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr16_+_23754772
|
4.813
|
NM_002738
NM_212535
|
PRKCB
|
protein kinase C, beta
|
|
chr4_-_40212670
|
4.804
|
NM_019027
|
RBM47
|
RNA binding motif protein 47
|
|
chr19_-_18369407
|
4.639
|
NM_145256
|
LRRC25
|
leucine rich repeat containing 25
|
|
chr12_+_91620749
|
4.599
|
NM_001037671
NM_001178097
|
C12orf74
|
chromosome 12 open reading frame 74
|
|
chr17_-_59363352
|
4.564
|
NM_000626
NM_001039933
NM_021602
|
CD79B
|
CD79b molecule, immunoglobulin-associated beta
|
|
chr5_-_176868483
|
4.563
|
|
DOK3
|
docking protein 3
|
|
chr10_-_98021144
|
4.353
|
NM_001114094
NM_013314
|
BLNK
|
B-cell linker
|
|
chr14_-_106119742
|
4.300
|
|
IGLJ3
IGHG1
|
immunoglobulin lambda joining 3
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr14_-_106064887
|
4.264
|
|
LOC100126583
IGHV3-48
IGHA1
IGHM
|
hypothetical LOC100126583
immunoglobulin heavy variable 3-48
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant mu
|
|
chr14_-_105862080
|
4.244
|
|
IGHV4-31
IGHA1
IGHD
IGHG1
IGHG2
IGHG3
IGHM
SCFV
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant delta
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 2 (G2m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
immunoglobulin heavy constant mu
single-chain Fv fragment
|
|
chr17_-_16816113
|
4.184
|
NM_012452
|
TNFRSF13B
|
tumor necrosis factor receptor superfamily, member 13B
|
|
chr22_+_20880159
|
3.983
|
|
IGLV6-57
|
immunoglobulin lambda variable 6-57
|
|
chr6_-_11887265
|
3.932
|
NM_001143948
NM_032744
|
C6orf105
|
chromosome 6 open reading frame 105
|
|
chr22_+_20316968
|
3.908
|
NM_152612
|
CCDC116
|
coiled-coil domain containing 116
|
|
chr22_+_21370394
|
3.803
|
|
IGL@
|
immunoglobulin lambda locus
|
|
chr9_+_94766379
|
3.706
|
|
FGD3
|
FYVE, RhoGEF and PH domain containing 3
|
|
chr17_-_33179118
|
3.688
|
NM_000458
NM_001165923
|
HNF1B
|
HNF1 homeobox B
|
|
chr11_+_527521
|
3.649
|
NM_198075
|
LRRC56
|
leucine rich repeat containing 56
|
|
chr19_+_17377502
|
3.465
|
|
LOC100507535
|
hypothetical LOC100507535
|
|
chr19_-_17377383
|
3.457
|
NM_004335
|
BST2
|
bone marrow stromal cell antigen 2
|
|
chr22_+_20880112
|
3.447
|
|
|
|
|
chr6_-_33016561
|
3.445
|
NM_002118
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta
|
|
chr1_+_205736095
|
3.432
|
NM_000573
NM_000651
|
CR1
|
complement component (3b/4b) receptor 1 (Knops blood group)
|
|
chr11_-_31789360
|
3.431
|
|
PAX6
|
paired box 6
|
|
chr14_-_105623358
|
3.225
|
|
IGHA1
IGHD
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant delta
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr9_-_139316523
|
3.147
|
NM_001004354
|
NRARP
|
NOTCH-regulated ankyrin repeat protein
|
|
chr17_-_33178987
|
3.138
|
|
HNF1B
|
HNF1 homeobox B
|
|
chr19_-_19142053
|
3.123
|
|
MEF2B
|
myocyte enhancer factor 2B
|
|
chr1_-_181889070
|
3.119
|
NM_203454
|
APOBEC4
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 4 (putative)
|
|
chr11_-_31789431
|
3.108
|
NM_000280
NM_001604
|
PAX6
|
paired box 6
|
|
chr19_-_19142090
|
3.105
|
NM_001145785
|
MEF2B
|
myocyte enhancer factor 2B
|
|
chr11_-_31789334
|
3.086
|
|
PAX6
|
paired box 6
|
|
chr1_+_200246370
|
3.046
|
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific )
|
|
chr16_-_73366200
|
3.040
|
NM_024306
|
FA2H
|
fatty acid 2-hydroxylase
|
|
chr6_-_138862273
|
3.013
|
|
|
|
|
chr16_-_73365975
|
2.993
|
|
FA2H
|
fatty acid 2-hydroxylase
|
|
chr14_-_105796665
|
2.972
|
|
IGHV4-31
IGHV3-23
IGHA1
IGHG1
IGHM
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy variable 3-23
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant mu
|
|
chr12_-_8656705
|
2.901
|
NM_020661
|
AICDA
|
activation-induced cytidine deaminase
|
|
chr19_+_14554895
|
2.898
|
NM_207390
|
CLEC17A
|
C-type lectin domain family 17, member A
|
|
chrX_-_24575146
|
2.892
|
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr22_-_22426551
|
2.861
|
NM_013378
|
VPREB3
|
pre-B lymphocyte 3
|
|
chr22_+_21340757
|
2.837
|
|
|
|
|
chr14_-_106202262
|
2.836
|
|
IGLJ3
IGHA1
|
immunoglobulin lambda joining 3
immunoglobulin heavy constant alpha 1
|
|
chr14_-_106241445
|
2.821
|
|
IGHV4-31
IGHV1-69
IGHA1
IGHG1
IGHG3
IGHM
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy variable 1-69
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
immunoglobulin heavy constant mu
|
|
chr6_+_34312667
|
2.803
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr1_+_200246312
|
2.778
|
NM_001114309
NM_004433
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific )
|
|
chr17_-_53849888
|
2.720
|
|
RNF43
|
ring finger protein 43
|
|
chrX_-_24575375
|
2.664
|
NM_001163265
NM_004845
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr6_+_53043729
|
2.663
|
NM_012347
|
FBXO9
|
F-box protein 9
|
|
chr22_+_21407066
|
2.616
|
|
|
|
|
chr10_+_124897627
|
2.579
|
NM_005519
|
HMX2
|
H6 family homeobox 2
|
|
chr14_-_105763152
|
2.548
|
|
IGHV4-31
IGHA1
IGHD
IGHG1
IGHG3
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant delta
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
|
|
chr6_-_25834733
|
2.528
|
NM_170745
|
HIST1H2AA
|
histone cluster 1, H2aa
|
|
chr11_-_31789168
|
2.508
|
|
PAX6
|
paired box 6
|
|
chr13_-_45640679
|
2.426
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr6_+_34312554
|
2.426
|
NM_002131
NM_145899
NM_145903
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr8_+_86287161
|
2.407
|
NM_001083589
|
E2F5
|
E2F transcription factor 5, p130-binding
|
|
chr22_+_21495273
|
2.402
|
|
|
|
|
chr3_-_52688768
|
2.356
|
NM_018165
NM_181042
|
PBRM1
|
polybromo 1
|
|
chr14_-_106184988
|
2.354
|
|
IGHA1
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr6_-_31668476
|
2.332
|
NM_001145466
NM_001145467
NM_147130
|
NCR3
|
natural cytotoxicity triggering receptor 3
|
|
chr17_-_8004671
|
2.327
|
|
VAMP2
|
vesicle-associated membrane protein 2 (synaptobrevin 2)
|
|
chr6_+_34312633
|
2.325
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr1_+_204797079
|
2.292
|
NM_182665
|
RASSF5
|
Ras association (RalGDS/AF-6) domain family member 5
|
|
chr6_+_34312641
|
2.290
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr21_+_38566298
|
2.289
|
NM_170737
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15
|
|
chr2_-_2313894
|
2.280
|
NM_015025
|
MYT1L
|
myelin transcription factor 1-like
|
|
chr6_+_34312625
|
2.278
|
NM_145901
NM_145902
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr14_-_106330781
|
2.271
|
|
IGHV5-78
|
immunoglobulin heavy variable 5-78 pseudogene
|
|
chr17_-_53849929
|
2.242
|
NM_017763
|
RNF43
|
ring finger protein 43
|
|
chr16_+_10878539
|
2.232
|
NM_000246
|
CIITA
|
class II, major histocompatibility complex, transactivator
|
|
chr19_-_18253048
|
2.156
|
|
JUND
|
jun D proto-oncogene
|
|
chr21_-_10120690
|
2.155
|
NM_001187
NM_182484
NM_181704
NM_182481
NM_182482
|
BAGE
BAGE5
BAGE4
BAGE3
BAGE2
|
B melanoma antigen
B melanoma antigen family, member 5
B melanoma antigen family, member 4
B melanoma antigen family, member 3
B melanoma antigen family, member 2
|
|
chr14_-_105796268
|
2.145
|
|
ZCWPW2
IGHV3-23
IGKV3-20
|
zinc finger, CW type with PWWP domain 2
immunoglobulin heavy variable 3-23
immunoglobulin kappa variable 3-20
|
|
chr15_-_38387357
|
2.131
|
|
PLCB2
|
phospholipase C, beta 2
|
|
chr2_-_2314087
|
2.080
|
|
MYT1L
|
myelin transcription factor 1-like
|
|
chr5_-_66528330
|
2.074
|
NM_005582
|
CD180
|
CD180 molecule
|
|
chr6_+_25834983
|
2.061
|
NM_170610
|
HIST1H2BA
|
histone cluster 1, H2ba
|
|
chr5_-_75044001
|
2.038
|
NM_152408
|
POC5
|
POC5 centriolar protein homolog (Chlamydomonas)
|
|
chr22_+_21384862
|
2.026
|
|
IGLV3-21
IGLJ3
IGLC1
|
immunoglobulin lambda variable 3-21
immunoglobulin lambda joining 3
immunoglobulin lambda constant 1 (Mcg marker)
|
|
chr5_+_60663842
|
2.026
|
NM_020928
|
ZSWIM6
|
zinc finger, SWIM-type containing 6
|
|
chr11_-_62125826
|
2.017
|
NM_004739
|
MTA2
|
metastasis associated 1 family, member 2
|
|
chr15_-_38387410
|
2.012
|
NM_004573
|
PLCB2
|
phospholipase C, beta 2
|
|
chr6_-_35764655
|
1.971
|
NM_001145776
NM_001145777
NM_004117
|
FKBP5
|
FK506 binding protein 5
|
|
chr12_+_56290229
|
1.951
|
NM_001111270
|
ARHGEF25
|
Rho guanine nucleotide exchange factor (GEF) 25
|
|
chr19_-_16111695
|
1.950
|
|
|
|
|
chr19_-_18253259
|
1.948
|
|
|
|
|
chr14_-_105762742
|
1.944
|
|
|
|
|
chr21_-_38415273
|
1.942
|
NM_005867
|
DSCR4
|
Down syndrome critical region gene 4
|
|
chr6_-_168219380
|
1.920
|
NM_001122841
|
FRMD1
|
FERM domain containing 1
|
|
chr21_+_29322100
|
1.878
|
|
USP16
|
ubiquitin specific peptidase 16
|
|
chr19_+_59820413
|
1.864
|
NM_006669
|
LILRB1
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 1
|
|
chr17_-_34575939
|
1.850
|
NM_001143968
|
ARL5C
|
ADP-ribosylation factor-like 5C
|
|
chr12_-_120270035
|
1.841
|
|
ANAPC5
|
anaphase promoting complex subunit 5
|
|
chr1_+_204797120
|
1.836
|
|
RASSF5
|
Ras association (RalGDS/AF-6) domain family member 5
|
|
chr3_+_120799411
|
1.797
|
NM_015900
|
PLA1A
|
phospholipase A1 member A
|
|
chr19_-_16111594
|
1.795
|
|
|
|
|
chr16_+_84202515
|
1.790
|
NM_001134473
|
KIAA0182
|
KIAA0182
|
|
chr6_+_34312979
|
1.789
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr20_-_48980858
|
1.763
|
NM_015339
NM_181442
|
ADNP
|
activity-dependent neuroprotector homeobox
|
|
chr6_+_43247179
|
1.724
|
|
SRF
|
serum response factor (c-fos serum response element-binding transcription factor)
|
|
chr5_-_131907071
|
1.710
|
NM_000879
|
IL5
|
interleukin 5 (colony-stimulating factor, eosinophil)
|
|
chrX_+_117410693
|
1.702
|
|
WDR44
|
WD repeat domain 44
|
|
chr17_+_70244553
|
1.702
|
NM_001163989
|
RAB37
|
RAB37, member RAS oncogene family
|
|
chr8_+_56954899
|
1.688
|
NM_001111097
NM_002350
|
LYN
|
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog
|
|
chrX_+_69591639
|
1.682
|
NM_001166278
|
DLG3
|
discs, large homolog 3 (Drosophila)
|
|
chr14_-_105681377
|
1.670
|
|
IGHV4-31
IGHG1
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr2_+_96790298
|
1.643
|
NM_020184
|
CNNM4
|
cyclin M4
|
|
chr6_+_34312906
|
1.622
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr19_+_3523950
|
1.621
|
|
HMG20B
|
high-mobility group 20B
|
|
chr14_-_105644300
|
1.618
|
|
IGHV4-31
IGHA1
IGHA2
IGHG1
IGHG3
IGHM
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant alpha 2 (A2m marker)
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
immunoglobulin heavy constant mu
|
|
chr6_-_35764588
|
1.614
|
|
FKBP5
|
FK506 binding protein 5
|
|
chr6_+_27323458
|
1.597
|
NM_005865
|
PRSS16
|
protease, serine, 16 (thymus)
|
|
chr2_-_238813284
|
1.591
|
|
HES6
|
hairy and enhancer of split 6 (Drosophila)
|
|
chr20_-_60415729
|
1.577
|
NM_031215
|
CABLES2
|
Cdk5 and Abl enzyme substrate 2
|
|
chr17_+_60563963
|
1.542
|
|
RGS9
|
regulator of G-protein signaling 9
|
|
chrX_+_123062143
|
1.533
|
|
STAG2
|
stromal antigen 2
|
|
chr6_+_34313007
|
1.526
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr21_-_35182842
|
1.510
|
NM_001001890
NM_001122607
|
RUNX1
|
runt-related transcription factor 1
|
|
chr6_-_10523203
|
1.503
|
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha)
|
|
chr6_+_26359805
|
1.501
|
NM_003524
|
HIST1H2BH
|
histone cluster 1, H2bh
|
|
chr6_-_138862271
|
1.493
|
NM_001144060
|
NHSL1
|
NHS-like 1
|
|
chr2_-_238813365
|
1.478
|
NM_001142853
NM_018645
|
HES6
|
hairy and enhancer of split 6 (Drosophila)
|
|
chr21_-_35182873
|
1.462
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr8_+_56954946
|
1.459
|
|
LYN
|
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog
|
|
chr17_+_60563917
|
1.425
|
NM_001081955
NM_001165933
NM_003835
|
RGS9
|
regulator of G-protein signaling 9
|
|
chrX_+_12903145
|
1.425
|
NM_021109
NM_183049
|
TMSB4X
TMSL3
|
thymosin beta 4, X-linked
thymosin-like 3
|
|
chr16_+_28850760
|
1.419
|
NM_001178098
NM_001770
|
CD19
|
CD19 molecule
|
|
chr11_-_34336102
|
1.412
|
NM_145804
|
ABTB2
|
ankyrin repeat and BTB (POZ) domain containing 2
|
|
chr19_-_18253413
|
1.403
|
NM_005354
|
JUND
|
jun D proto-oncogene
|
|
chr14_-_105785639
|
1.403
|
|
|
|
|
chr19_+_3523958
|
1.375
|
|
HMG20B
|
high-mobility group 20B
|
|
chr15_-_38953628
|
1.368
|
NM_133639
|
RHOV
|
ras homolog gene family, member V
|
|
chr6_+_43247168
|
1.367
|
|
SRF
|
serum response factor (c-fos serum response element-binding transcription factor)
|
|
chr6_+_37511383
|
1.366
|
|
|
|
|
chr3_-_46043982
|
1.363
|
NM_001024644
NM_005283
|
XCR1
|
chemokine (C motif) receptor 1
|
|
chr17_-_71287417
|
1.354
|
NM_005324
|
H3F3B
|
H3 histone, family 3B (H3.3B)
|
|
chr8_+_19841120
|
1.352
|
|
LPL
|
lipoprotein lipase
|
|
chrX_+_37524250
|
1.337
|
|
CYBB
|
cytochrome b-245, beta polypeptide
|
|
chr22_+_21042091
|
1.336
|
|
IGLV1-44
IGL@
|
immunoglobulin lambda variable 1-44
immunoglobulin lambda locus
|
|
chr16_+_2959362
|
1.330
|
|
PAQR4
|
progestin and adipoQ receptor family member IV
|
|
chr6_+_135558572
|
1.327
|
|
|
|
|
chr8_+_7789608
|
1.323
|
NM_004942
|
DEFB4A
|
defensin, beta 4A
|
|
chr14_-_105524207
|
1.319
|
|
IGHG1
|
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr22_+_44059491
|
1.318
|
NM_001167574
NM_006953
|
UPK3A
|
uroplakin 3A
|
|
chr19_-_18253436
|
1.317
|
|
JUND
|
jun D proto-oncogene
|
|
chrX_+_37524190
|
1.286
|
NM_000397
|
CYBB
|
cytochrome b-245, beta polypeptide
|
|
chr1_+_156526182
|
1.270
|
NM_001765
|
CD1C
|
CD1c molecule
|
|
chr17_+_70495318
|
1.265
|
NM_014603
|
CDR2L
|
cerebellar degeneration-related protein 2-like
|
|
chr8_+_19840861
|
1.257
|
NM_000237
|
LPL
|
lipoprotein lipase
|
|
chr6_+_32515596
|
1.254
|
NM_019111
|
HLA-DRA
|
major histocompatibility complex, class II, DR alpha
|
|
chr3_-_3127057
|
1.237
|
NM_000564
NM_175724
NM_175725
NM_175726
NM_175727
NM_175728
|
IL5RA
|
interleukin 5 receptor, alpha
|
|
chr4_-_40211324
|
1.216
|
|
RBM47
|
RNA binding motif protein 47
|
|
chr1_+_5975345
|
1.210
|
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chr8_+_19841010
|
1.199
|
|
LPL
|
lipoprotein lipase
|
|
chr7_-_36730525
|
1.197
|
NM_001177506
NM_001177507
NM_001637
|
AOAH
|
acyloxyacyl hydrolase (neutrophil)
|
|
chr17_+_77910411
|
1.187
|
NM_207459
|
TEX19
|
testis expressed 19
|
|
chr22_+_20899155
|
1.183
|
|
|
|
|
chr14_-_105589467
|
1.179
|
|
LOC100126583
IGHA1
IGHG1
IGHG2
IGHM
|
hypothetical LOC100126583
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 2 (G2m marker)
immunoglobulin heavy constant mu
|
|
chr7_+_94130673
|
1.165
|
|
PEG10
|
paternally expressed 10
|
|
chr16_+_2959342
|
1.165
|
NM_152341
|
PAQR4
|
progestin and adipoQ receptor family member IV
|
|
chr2_-_113310796
|
1.163
|
NM_000576
|
IL1B
|
interleukin 1, beta
|
|
chr1_+_226712430
|
1.159
|
NM_175055
|
HIST3H2BB
|
histone cluster 3, H2bb
|
|
chr4_-_71751070
|
1.157
|
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides
|
|
chr7_+_26404711
|
1.150
|
|
LOC441204
|
hypothetical locus LOC441204
|
|
chr4_-_91979113
|
1.146
|
NM_183049
|
TMSL3
|
thymosin-like 3
|
|
chr12_-_11030777
|
1.143
|
NM_176890
|
TAS2R50
|
taste receptor, type 2, member 50
|
|
chr10_+_97505398
|
1.140
|
NM_001164178
NM_001164181
NM_001164179
NM_001164182
NM_001164183
NM_001776
|
ENTPD1
|
ectonucleoside triphosphate diphosphohydrolase 1
|
|
chr12_-_55614311
|
1.139
|
NM_148897
|
SDR9C7
|
short chain dehydrogenase/reductase family 9C, member 7
|
|
chr6_-_53121540
|
1.115
|
NM_003643
|
GCM1
|
glial cells missing homolog 1 (Drosophila)
|
|
chr19_-_50779924
|
1.112
|
NM_001017989
NM_025136
|
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia)
|
|
chr14_-_106282128
|
1.087
|
|
IGHG1
|
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr3_+_122794859
|
1.065
|
NM_016298
|
FBXO40
|
F-box protein 40
|
|
chr22_+_21065207
|
1.063
|
|
IGLJ3
|
immunoglobulin lambda joining 3
|
|
chr12_-_48902692
|
1.057
|
NM_001113547
|
LIMA1
|
LIM domain and actin binding 1
|
|
chr2_+_173794692
|
1.050
|
|
ZAK
|
sterile alpha motif and leucine zipper containing kinase AZK
|
|
chr17_+_7494964
|
1.038
|
NM_001678
|
ATP1B2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide
|
|
chr22_+_21065202
|
1.034
|
|
|
|
|
chr11_-_62125789
|
1.027
|
|
MTA2
|
metastasis associated 1 family, member 2
|
|
chr14_-_105542441
|
1.026
|
|
IGLJ3
IGHA1
IGHG1
IGHM
|
immunoglobulin lambda joining 3
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant mu
|
|
chr6_-_26324850
|
1.022
|
NM_003518
|
HIST1H2BG
NCALD
|
histone cluster 1, H2bg
neurocalcin delta
|
|
chr19_-_50779914
|
1.018
|
|
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia)
|
|
chr22_+_21065134
|
1.016
|
|
LOC100290481
|
immunoglobulin lambda light chain-like
|
|
chr14_-_106270497
|
1.010
|
|
IGHG3
|
immunoglobulin heavy constant gamma 3 (G3m marker)
|
|
chr7_+_142629251
|
1.007
|
NM_176882
|
TAS2R40
|
taste receptor, type 2, member 40
|
|
chr11_-_83071084
|
1.007
|
NM_001142702
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr17_+_7495140
|
0.997
|
|
ATP1B2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide
|