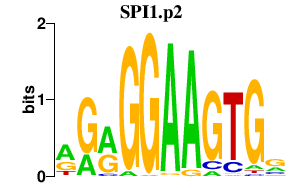
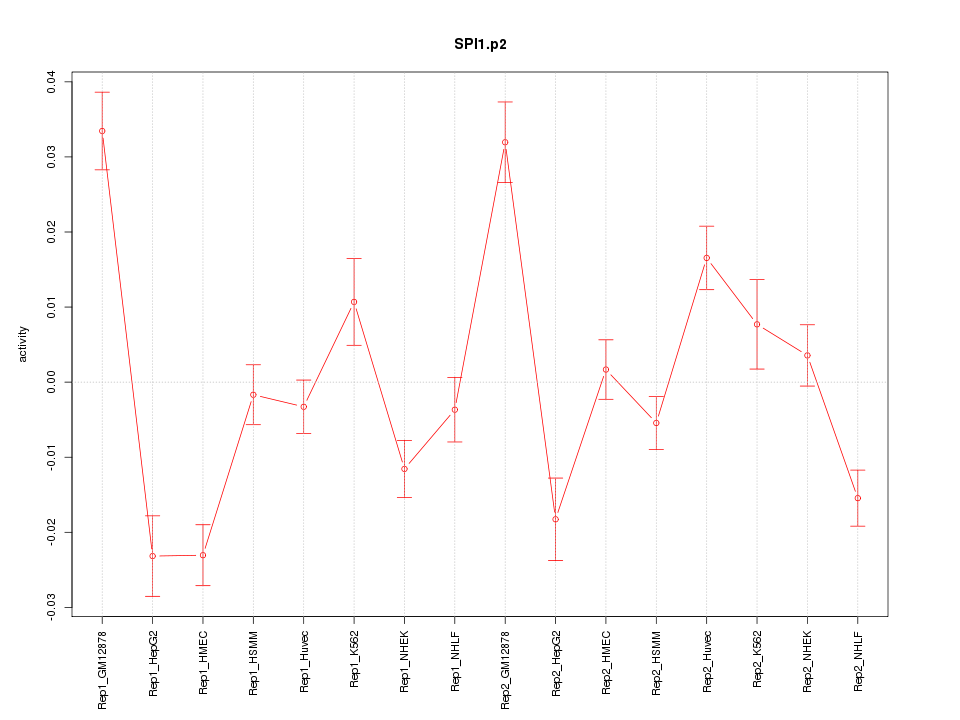
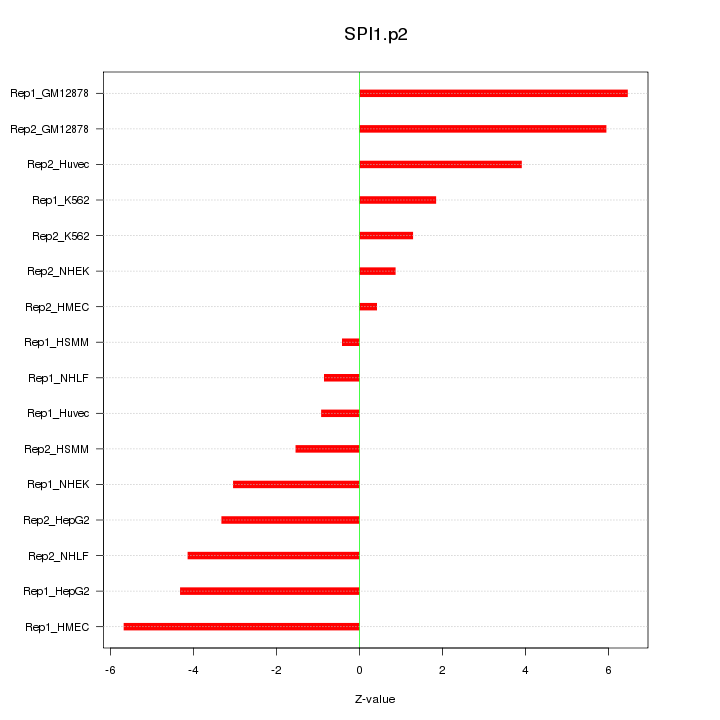
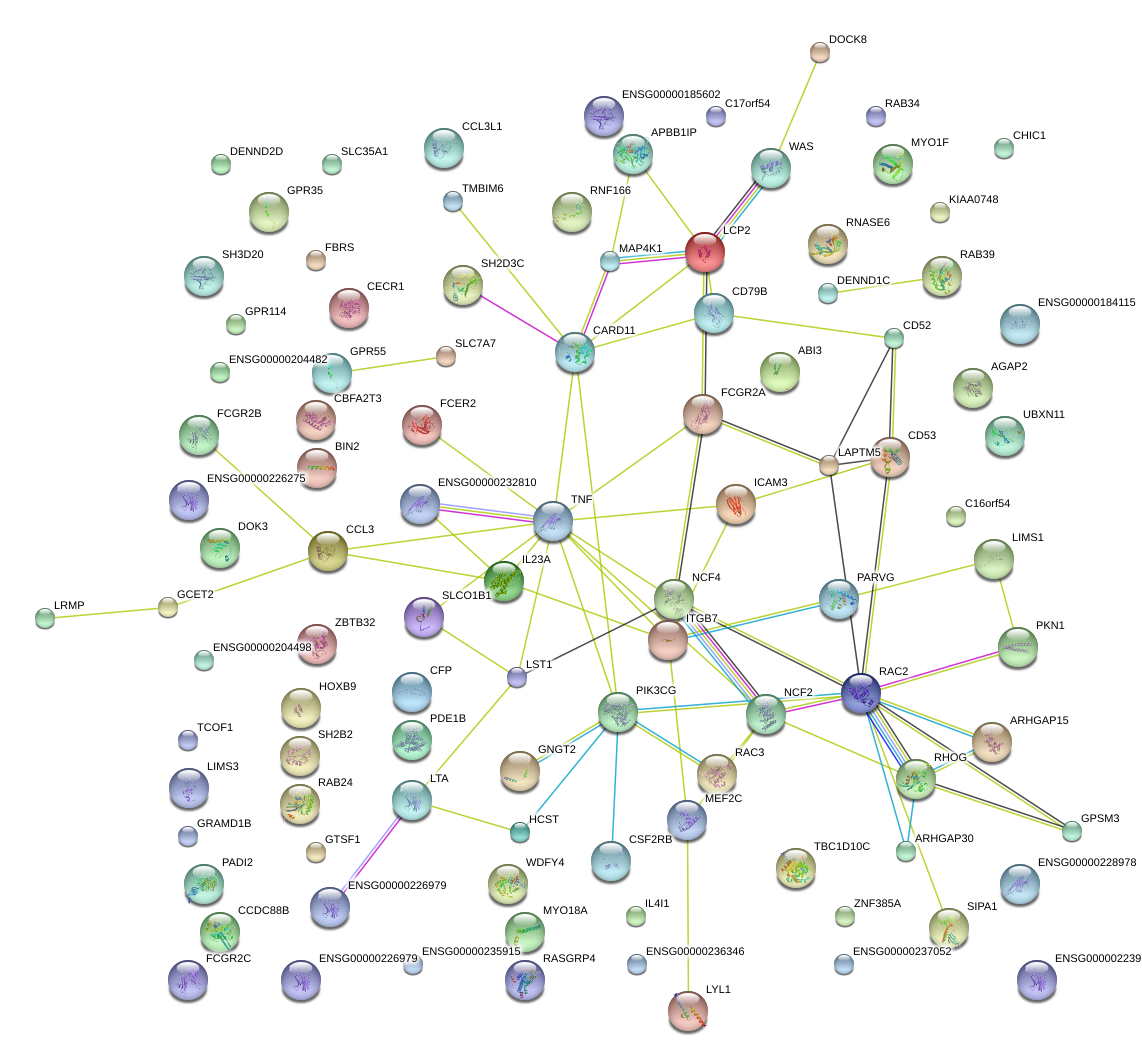
|
chr22_+_35586950
|
6.694
|
NM_000631
NM_013416
|
NCF4
|
neutrophil cytosolic factor 4, 40kDa
|
|
chr16_-_87570716
|
6.292
|
NM_005187
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chr11_+_63864263
|
5.869
|
NM_032251
|
CCDC88B
|
coiled-coil domain containing 88B
|
|
chr1_+_159899528
|
5.332
|
NM_001002273
NM_001002274
NM_001002275
NM_001190828
NM_004001
|
FCGR2B
FCGR2C
|
Fc fragment of IgG, low affinity IIb, receptor (CD32)
Fc fragment of IgG, low affinity IIc, receptor for (CD32) (gene/pseudogene)
|
|
chr10_+_49562866
|
4.628
|
|
WDFY4
|
WDFY family member 4
|
|
chrX_-_47374187
|
4.397
|
NM_001145252
|
CFP
|
complement factor properdin
|
|
chr11_+_65164767
|
4.253
|
|
SIPA1
|
signal-induced proliferation-associated 1
|
|
chr16_-_29664765
|
4.140
|
NM_175900
|
C16orf54
|
chromosome 16 open reading frame 54
|
|
chr12_-_55929189
|
4.038
|
|
|
|
|
chr1_-_26517342
|
3.895
|
NM_145345
|
UBXN11
|
UBX domain protein 11
|
|
chr1_+_159817752
|
3.865
|
NM_201563
|
FCGR2C
|
Fc fragment of IgG, low affinity IIc, receptor for (CD32) (gene/pseudogene)
|
|
chr6_-_32268267
|
3.706
|
|
GPSM3
|
G-protein signaling modulator 3
|
|
chr17_-_44058612
|
3.479
|
NM_024017
|
HOXB9
|
homeobox B9
|
|
chr1_+_26517032
|
3.353
|
|
CD52
|
CD52 molecule
|
|
chr7_+_106292932
|
3.286
|
|
PIK3CG
|
phosphoinositide-3-kinase, catalytic, gamma polypeptide
|
|
chr7_+_106292975
|
3.278
|
|
PIK3CG
|
phosphoinositide-3-kinase, catalytic, gamma polypeptide
|
|
chr12_-_53071285
|
3.165
|
NM_001130967
NM_001130968
|
ZNF385A
|
zinc finger protein 385A
|
|
chr17_+_44643067
|
3.159
|
|
ABI3
|
ABI family, member 3
|
|
chr1_+_26517045
|
3.154
|
|
CD52
|
CD52 molecule
|
|
chr19_-_10311262
|
3.118
|
NM_002162
|
ICAM3
|
intercellular adhesion molecule 3
|
|
chrX_-_47374647
|
3.067
|
NM_002621
|
CFP
|
complement factor properdin
|
|
chr17_-_40843530
|
3.059
|
|
ARHGAP27
|
Rho GTPase activating protein 27
|
|
chr17_+_44642587
|
2.979
|
NM_001135186
NM_016428
|
ABI3
|
ABI family, member 3
|
|
chr14_+_20319049
|
2.918
|
NM_005615
|
RNASE6
|
ribonuclease, RNase A family, k6
|
|
chr16_+_28413421
|
2.850
|
NM_018690
|
APOBR
|
apolipoprotein B receptor
|
|
chr17_-_31441390
|
2.843
|
NM_002983
|
CCL3
|
chemokine (C-C motif) ligand 3
|
|
chr7_+_101715110
|
2.760
|
NM_020979
|
SH2B2
|
SH2B adaptor protein 2
|
|
chr1_-_31003260
|
2.758
|
NM_006762
|
LAPTM5
|
lysosomal protein transmembrane 5
|
|
chr6_-_32268615
|
2.735
|
|
GPSM3
|
G-protein signaling modulator 3
|
|
chr14_-_22354457
|
2.712
|
|
SLC7A7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7
|
|
chr19_-_7672978
|
2.708
|
NM_002002
|
FCER2
|
Fc fragment of IgE, low affinity II, receptor for (CD23)
|
|
chr7_+_101715137
|
2.650
|
|
SH2B2
|
SH2B adaptor protein 2
|
|
chr12_-_51887266
|
2.632
|
NM_000889
|
ITGB7
|
integrin, beta 7
|
|
chr12_-_53664722
|
2.606
|
NM_001136030
|
KIAA0748
|
KIAA0748
|
|
chr1_+_111544469
|
2.597
|
|
|
|
|
chr6_+_31648071
|
2.579
|
NM_000595
|
LTA
|
lymphotoxin alpha (TNF superfamily, member 1)
|
|
chr16_+_56134077
|
2.555
|
NM_153837
|
GPR114
|
G protein-coupled receptor 114
|
|
chr16_+_56134037
|
2.421
|
|
GPR114
|
G protein-coupled receptor 114
|
|
chr2_+_143603439
|
2.421
|
|
ARHGAP15
|
Rho GTPase activating protein 15
|
|
chr14_-_22354389
|
2.419
|
|
SLC7A7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7
|
|
chrX_+_48427103
|
2.414
|
NM_000377
|
WAS
|
Wiskott-Aldrich syndrome (eczema-thrombocytopenia)
|
|
chrX_+_48427137
|
2.411
|
|
WAS
|
Wiskott-Aldrich syndrome (eczema-thrombocytopenia)
|
|
chr22_+_42908612
|
2.407
|
|
PARVG
|
parvin, gamma
|
|
chr1_-_111544603
|
2.388
|
|
|
|
|
chr2_+_108604111
|
2.383
|
NM_001193484
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr12_+_25096472
|
2.358
|
NM_006152
|
LRMP
|
lymphoid-restricted membrane protein
|
|
chr9_+_263025
|
2.357
|
NM_001190458
NM_001193536
|
DOCK8
|
dedicator of cytokinesis 8
|
|
chr19_+_14412052
|
2.342
|
|
PKN1
|
protein kinase N1
|
|
chr19_+_14412087
|
2.325
|
|
PKN1
|
protein kinase N1
|
|
chr12_-_50004102
|
2.315
|
|
BIN2
|
bridging integrator 2
|
|
chr19_+_14412080
|
2.314
|
NM_213560
|
PKN1
|
protein kinase N1
|
|
chr2_+_143603368
|
2.311
|
NM_018460
|
ARHGAP15
|
Rho GTPase activating protein 15
|
|
chr11_+_66927976
|
2.291
|
NM_198517
|
TBC1D10C
|
TBC1 domain family, member 10C
|
|
chr19_-_55091942
|
2.270
|
NM_152899
|
IL4I1
|
interleukin 4 induced 1
|
|
chrX_+_72699834
|
2.241
|
|
CHIC1
|
cysteine-rich hydrophobic domain 1
|
|
chr12_-_50004172
|
2.230
|
NM_016293
|
BIN2
|
bridging integrator 2
|
|
chr3_-_113334657
|
2.212
|
NM_001190259
NM_001190260
NM_152785
|
GCET2
|
germinal center expressed transcript 2
|
|
chr12_-_50004208
|
2.208
|
|
BIN2
|
bridging integrator 2
|
|
chr1_-_111544784
|
2.203
|
NM_024901
|
DENND2D
|
DENN/MADD domain containing 2D
|
|
chr17_-_24491521
|
2.163
|
|
MYO18A
|
myosin XVIIIA
|
|
chr12_+_48443232
|
2.121
|
|
TMBIM6
|
transmembrane BAX inhibitor motif containing 6
|
|
chr1_+_111217314
|
2.104
|
|
CD53
|
CD53 molecule
|
|
chr22_+_35639564
|
2.104
|
NM_000395
|
CSF2RB
|
colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage)
|
|
chr6_+_31647854
|
2.100
|
NM_001159740
|
LTA
|
lymphotoxin alpha (TNF superfamily, member 1)
|
|
chr1_-_111544820
|
2.099
|
|
DENND2D
|
DENN/MADD domain containing 2D
|
|
chr11_+_66927955
|
2.095
|
|
TBC1D10C
|
TBC1 domain family, member 10C
|
|
chr2_-_231498184
|
2.091
|
NM_005683
|
GPR55
|
G protein-coupled receptor 55
|
|
chr1_+_111217244
|
2.077
|
NM_000560
|
CD53
|
CD53 molecule
|
|
chr19_-_6432765
|
2.061
|
NM_024898
|
DENND1C
|
DENN/MADD domain containing 1C
|
|
chr6_+_31661934
|
2.038
|
NM_205838
NM_205839
|
LST1
|
leukocyte specific transcript 1
|
|
chr6_+_31662454
|
2.037
|
NM_205837
|
LST1
|
leukocyte specific transcript 1
|
|
chr2_+_241213334
|
1.961
|
NM_005301
|
GPR35
|
G protein-coupled receptor 35
|
|
chr12_+_53241365
|
1.937
|
NM_001165975
|
PDE1B
|
phosphodiesterase 1B, calmodulin-dependent
|
|
chr12_-_51887318
|
1.932
|
|
ITGB7
|
integrin, beta 7
|
|
chr16_-_87297525
|
1.920
|
NM_001171816
|
RNF166
|
ring finger protein 166
|
|
chr11_+_107304344
|
1.908
|
NM_017516
|
RAB39
|
RAB39, member RAS oncogene family
|
|
chr10_+_26767698
|
1.907
|
|
APBB1IP
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein
|
|
chr12_-_53153613
|
1.895
|
|
GTSF1
|
gametocyte specific factor 1
|
|
chr1_-_17318452
|
1.869
|
NM_007365
|
PADI2
|
peptidyl arginine deiminase, type II
|
|
chr16_+_30583278
|
1.868
|
NM_001105079
|
FBRS
|
fibrosin
|
|
chr12_-_53153639
|
1.867
|
NM_144594
|
GTSF1
|
gametocyte specific factor 1
|
|
chr7_-_3049872
|
1.866
|
NM_032415
|
CARD11
|
caspase recruitment domain family, member 11
|
|
chr19_-_13074973
|
1.865
|
NM_005583
|
LYL1
|
lymphoblastic leukemia derived sequence 1
|
|
chr1_-_181826292
|
1.859
|
NM_000433
NM_001190789
NM_001190794
|
NCF2
|
neutrophil cytosolic factor 2
|
|
chr19_-_43800374
|
1.854
|
NM_001042600
NM_007181
|
MAP4K1
|
mitogen-activated protein kinase kinase kinase kinase 1
|
|
chr9_-_129580732
|
1.853
|
NM_170600
|
SH2D3C
|
SH2 domain containing 3C
|
|
chr17_-_44642934
|
1.852
|
NM_001198754
|
GNGT2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2
|
|
chr5_-_88215034
|
1.816
|
NM_001193350
NM_002397
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr19_-_8548297
|
1.813
|
|
MYO1F
|
myosin IF
|
|
chr19_-_43608641
|
1.805
|
NM_001146202
NM_001146203
NM_001146204
NM_001146205
NM_001146206
NM_001146207
NM_170604
|
RASGRP4
|
RAS guanyl releasing protein 4
|
|
chr5_+_140871107
|
1.804
|
|
|
|
|
chr19_+_40895669
|
1.802
|
NM_014383
|
ZBTB32
|
zinc finger and BTB domain containing 32
|
|
chr5_-_169657192
|
1.801
|
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr22_-_35970230
|
1.801
|
NM_002872
|
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2)
|
|
chr19_-_8548232
|
1.798
|
|
MYO1F
|
myosin IF
|
|
chr22_-_16080265
|
1.795
|
|
CECR1
|
cat eye syndrome chromosome region, candidate 1
|
|
chr17_-_59363352
|
1.794
|
NM_000626
NM_001039933
NM_021602
|
CD79B
|
CD79b molecule, immunoglobulin-associated beta
|
|
chr19_+_41085221
|
1.785
|
NM_001007469
NM_014266
|
HCST
|
hematopoietic cell signal transducer
|
|
chr11_+_65164167
|
1.781
|
NM_153253
|
SIPA1
|
signal-induced proliferation-associated 1
|
|
chr12_-_56422120
|
1.773
|
NM_014770
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2
|
|
chr7_+_106293112
|
1.740
|
NM_002649
|
PIK3CG
|
phosphoinositide-3-kinase, catalytic, gamma polypeptide
|
|
chr11_+_122936246
|
1.734
|
|
GRAMD1B
|
GRAM domain containing 1B
|
|
chr6_+_31651319
|
1.731
|
NM_000594
|
TNF
|
tumor necrosis factor
|
|
chr12_+_55018925
|
1.729
|
NM_016584
|
IL23A
|
interleukin 23, alpha subunit p19
|
|
chr1_-_159306037
|
1.727
|
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr19_-_13074669
|
1.719
|
|
LYL1
|
lymphoblastic leukemia derived sequence 1
|
|
chr5_-_176869440
|
1.711
|
|
DOK3
|
docking protein 3
|
|
chrX_-_153632355
|
1.704
|
NM_001081573
NM_080612
|
GAB3
|
GRB2-associated binding protein 3
|
|
chr19_-_46504831
|
1.701
|
|
|
|
|
chr5_-_169657312
|
1.701
|
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr16_+_29582065
|
1.699
|
|
SPN
|
sialophorin
|
|
chr16_+_29582069
|
1.697
|
NM_003123
|
SPN
|
sialophorin
|
|
chr11_-_66961709
|
1.690
|
NM_005608
|
PTPRCAP
|
protein tyrosine phosphatase, receptor type, C-associated protein
|
|
chr7_+_73826244
|
1.679
|
NM_000265
|
NCF1
NCF1B
NCF1C
|
neutrophil cytosolic factor 1
neutrophil cytosolic factor 1B pseudogene
neutrophil cytosolic factor 1C pseudogene
|
|
chr6_-_33016561
|
1.678
|
NM_002118
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta
|
|
chr17_+_31455332
|
1.667
|
NM_002984
|
CCL4
|
chemokine (C-C motif) ligand 4
|
|
chr22_-_35970133
|
1.665
|
|
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2)
|
|
chr19_-_15436333
|
1.663
|
NM_022904
|
RASAL3
|
RAS protein activator like 3
|
|
chr1_-_31003181
|
1.662
|
|
LAPTM5
|
lysosomal protein transmembrane 5
|
|
chr9_-_129707392
|
1.659
|
|
LOC100131355
ST6GALNAC6
|
hypothetical protein LOC100131355
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6
|
|
chr1_+_207995999
|
1.638
|
NM_025228
|
TRAF3IP3
|
TRAF3 interacting protein 3
|
|
chr6_+_43846735
|
1.627
|
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr2_-_175207521
|
1.623
|
NM_003387
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr5_-_169657360
|
1.621
|
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr2_+_68445936
|
1.619
|
|
PLEK
|
pleckstrin
|
|
chr5_-_169657395
|
1.612
|
NM_005565
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr19_+_59866217
|
1.610
|
|
LILRB4
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 4
|
|
chr1_-_181826086
|
1.588
|
|
NCF2
|
neutrophil cytosolic factor 2
|
|
chr3_+_195889845
|
1.588
|
|
FAM43A
|
family with sequence similarity 43, member A
|
|
chr2_+_68445821
|
1.587
|
NM_002664
|
PLEK
|
pleckstrin
|
|
chr1_+_26516967
|
1.582
|
NM_001803
|
CD52
|
CD52 molecule
|
|
chr16_+_48865521
|
1.581
|
|
ADCY7
|
adenylate cyclase 7
|
|
chr1_-_92124064
|
1.577
|
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr1_-_21375939
|
1.576
|
NM_001198801
NM_001198802
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3
|
|
chr6_+_37511383
|
1.572
|
|
|
|
|
chr19_+_54530497
|
1.554
|
|
CD37
|
CD37 molecule
|
|
chr22_+_23153529
|
1.547
|
NM_000675
|
ADORA2A
|
adenosine A2a receptor
|
|
chr12_-_53065758
|
1.546
|
|
ZNF385A
|
zinc finger protein 385A
|
|
chr22_-_28992787
|
1.544
|
NM_020530
|
OSM
|
oncostatin M
|
|
chr5_-_176869943
|
1.536
|
NM_001144875
NM_001144876
|
DOK3
|
docking protein 3
|
|
chr1_-_158883666
|
1.535
|
NM_003037
|
SLAMF1
|
signaling lymphocytic activation molecule family member 1
|
|
chr12_+_25096755
|
1.524
|
|
LRMP
|
lymphoid-restricted membrane protein
|
|
chr12_-_56159631
|
1.523
|
NM_001080157
|
ARHGAP9
|
Rho GTPase activating protein 9
|
|
chr8_+_145158432
|
1.509
|
NM_001134374
NM_198572
|
SPATC1
|
spermatogenesis and centriole associated 1
|
|
chr5_-_88016397
|
1.499
|
|
LOC645323
|
hypothetical LOC645323
|
|
chr22_-_36212160
|
1.497
|
NM_001166343
NM_002405
|
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr8_+_126511739
|
1.478
|
NM_025195
|
TRIB1
|
tribbles homolog 1 (Drosophila)
|
|
chr11_+_46339691
|
1.473
|
NM_001105540
|
DGKZ
|
diacylglycerol kinase, zeta
|
|
chr2_+_169637255
|
1.471
|
NM_001142270
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9
|
|
chr11_+_122936145
|
1.465
|
|
GRAMD1B
|
GRAM domain containing 1B
|
|
chr7_-_4867963
|
1.461
|
NM_020144
|
PAPOLB
|
poly(A) polymerase beta (testis specific)
|
|
chr19_-_40925151
|
1.446
|
NM_024660
|
TMEM149
|
transmembrane protein 149
|
|
chr4_+_91035025
|
1.444
|
NM_007351
|
MMRN1
|
multimerin 1
|
|
chr17_-_35975227
|
1.437
|
NM_001838
|
CCR7
|
chemokine (C-C motif) receptor 7
|
|
chr19_-_55008172
|
1.434
|
NM_001171937
NM_025129
|
FUZ
|
fuzzy homolog (Drosophila)
|
|
chr15_-_76313896
|
1.434
|
NM_015162
|
ACSBG1
|
acyl-CoA synthetase bubblegum family member 1
|
|
chr10_-_75080784
|
1.419
|
NM_024875
|
SYNPO2L
|
synaptopodin 2-like
|
|
chr14_-_22354851
|
1.409
|
NM_001126105
NM_003982
|
SLC7A7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7
|
|
chr12_+_6930815
|
1.408
|
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6
|
|
chr10_+_26767657
|
1.404
|
|
APBB1IP
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein
|
|
chr3_-_114176401
|
1.403
|
NM_138806
NM_138939
NM_138940
NM_170780
|
CD200R1
|
CD200 receptor 1
|
|
chr19_-_55104875
|
1.400
|
|
NUP62
|
nucleoporin 62kDa
|
|
chr16_-_30301634
|
1.398
|
NM_052838
|
SEPT1
|
septin 1
|
|
chr21_-_45165204
|
1.393
|
|
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit)
|
|
chr21_-_45165302
|
1.382
|
NM_000211
|
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit)
|
|
chr16_+_27320983
|
1.381
|
NM_181078
NM_181079
|
IL21R
|
interleukin 21 receptor
|
|
chr17_+_53769562
|
1.374
|
|
LOC100506779
|
hypothetical LOC100506779
|
|
chr22_-_30071761
|
1.374
|
|
PATZ1
|
POZ (BTB) and AT hook containing zinc finger 1
|
|
chr22_+_23333610
|
1.373
|
NM_001032365
|
GGT1
|
gamma-glutamyltransferase 1
|
|
chr7_+_149895390
|
1.373
|
NM_018326
|
GIMAP4
|
GTPase, IMAP family member 4
|
|
chr10_-_71002998
|
1.371
|
NM_020999
|
NEUROG3
|
neurogenin 3
|
|
chr1_-_158815895
|
1.370
|
NM_001184879
NM_001184881
NM_001184882
NM_003874
|
CD84
|
CD84 molecule
|
|
chr6_+_37081387
|
1.367
|
NM_173558
|
FGD2
|
FYVE, RhoGEF and PH domain containing 2
|
|
chr16_+_29582480
|
1.360
|
|
SPN
|
sialophorin
|
|
chr20_+_30019465
|
1.357
|
NM_001011718
|
XKR7
|
XK, Kell blood group complex subunit-related family, member 7
|
|
chr21_-_45165249
|
1.354
|
|
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit)
|
|
chr1_+_149309703
|
1.352
|
NM_144618
|
GABPB2
|
GA binding protein transcription factor, beta subunit 2
|
|
chr1_-_205161821
|
1.343
|
NM_001193338
NM_001142473
NM_005449
|
FAIM3
|
Fas apoptotic inhibitory molecule 3
|
|
chr3_-_122862447
|
1.339
|
NM_005335
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr17_+_53769659
|
1.321
|
|
LOC100506779
|
hypothetical LOC100506779
|
|
chr12_-_53661888
|
1.320
|
NM_001098815
|
KIAA0748
|
KIAA0748
|
|
chr3_-_122862391
|
1.318
|
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr22_+_42908548
|
1.300
|
|
PARVG
|
parvin, gamma
|
|
chr1_-_158815850
|
1.291
|
|
CD84
|
CD84 molecule
|
|
chr11_-_46099531
|
1.288
|
NM_001101802
NM_016621
|
PHF21A
|
PHD finger protein 21A
|
|
chr22_+_42908589
|
1.287
|
NM_001137606
|
PARVG
|
parvin, gamma
|
|
chr7_-_81237162
|
1.281
|
NM_000601
NM_001010931
NM_001010932
NM_001010933
NM_001010934
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor)
|
|
chr7_+_150065376
|
1.280
|
NM_018384
|
GIMAP5
|
GTPase, IMAP family member 5
|
|
chr5_-_142795269
|
1.275
|
NM_001018074
NM_001018075
NM_001018077
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor)
|
|
chrX_-_70248022
|
1.271
|
NM_000206
|
IL2RG
|
interleukin 2 receptor, gamma
|
|
chr1_+_154386358
|
1.271
|
NM_001193301
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A
|
|
chr7_+_149779011
|
1.266
|
|
GIMAP8
|
GTPase, IMAP family member 8
|
|
chr1_-_159306327
|
1.266
|
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr7_+_128367961
|
1.266
|
NM_001098627
|
IRF5
|
interferon regulatory factor 5
|
|
chr12_-_56159895
|
1.253
|
NM_032496
|
ARHGAP9
|
Rho GTPase activating protein 9
|
|
chr6_+_73388713
|
1.245
|
|
KCNQ5
|
potassium voltage-gated channel, KQT-like subfamily, member 5
|
|
chr6_-_6265896
|
1.241
|
NM_000129
|
F13A1
|
coagulation factor XIII, A1 polypeptide
|
|
chr7_-_44985202
|
1.230
|
NM_033054
|
MYO1G
|
myosin IG
|
|
chr12_-_8656705
|
1.229
|
NM_020661
|
AICDA
|
activation-induced cytidine deaminase
|
|
chr5_+_88220846
|
1.226
|
|
|
|