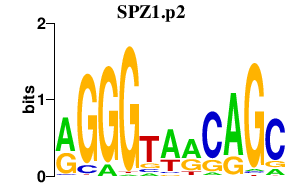
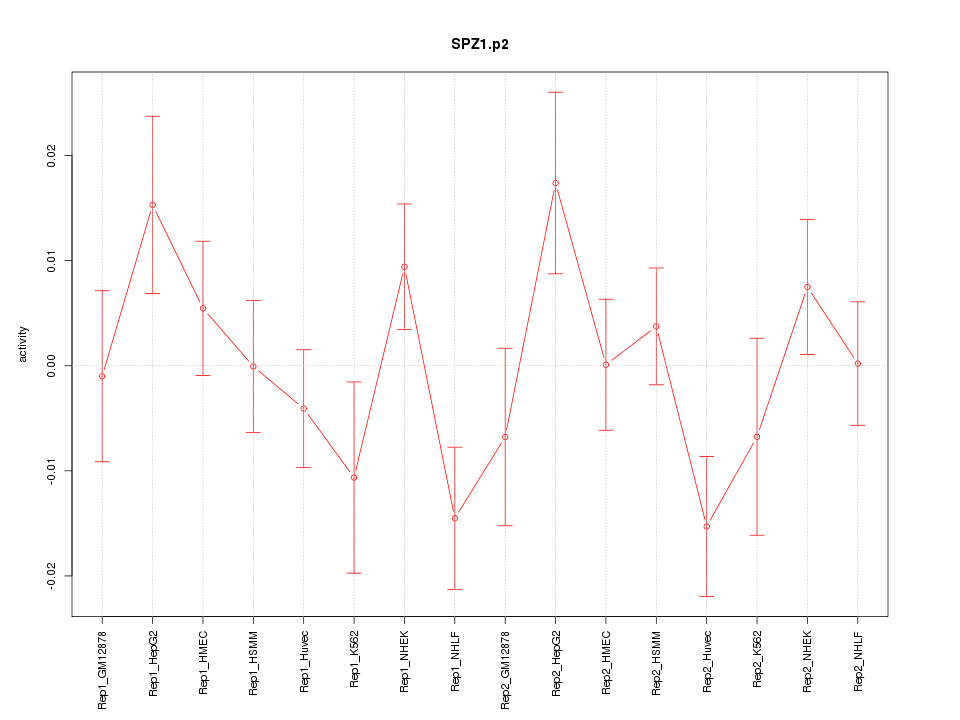
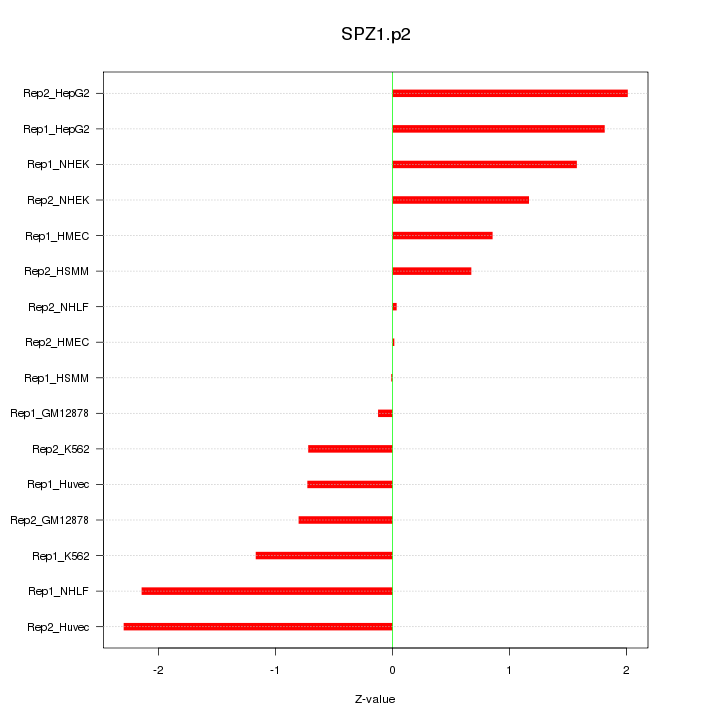
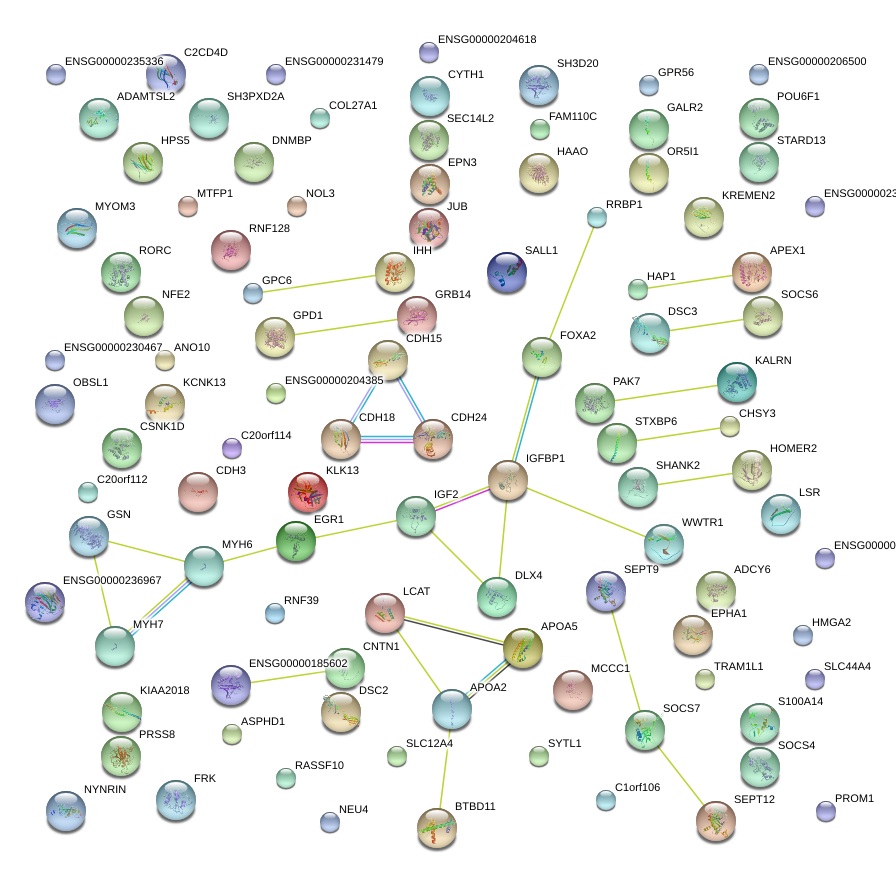
|
chr1_+_199130571
|
1.309
|
NM_001142569
|
C1orf106
|
chromosome 1 open reading frame 106
|
|
chr16_-_66535457
|
1.223
|
NM_000229
|
LCAT
|
lecithin-cholesterol acyltransferase
|
|
chr17_+_72795567
|
1.151
|
NM_001113492
|
SEPT9
|
septin 9
|
|
chr6_-_116488458
|
1.135
|
NM_002031
|
FRK
|
fyn-related kinase
|
|
chrX_+_105856531
|
1.097
|
NM_194463
|
RNF128
|
ring finger protein 128
|
|
chr2_-_36384
|
1.025
|
NM_001077710
|
FAM110C
|
family with sequence similarity 110, member C
|
|
chr16_+_29819647
|
0.975
|
NM_181718
|
ASPHD1
|
aspartate beta-hydroxylase domain containing 1
|
|
chr20_-_30535045
|
0.935
|
|
C20orf112
|
chromosome 20 open reading frame 112
|
|
chr2_-_220144254
|
0.922
|
|
OBSL1
|
obscurin-like 1
|
|
chr10_-_101680645
|
0.905
|
|
DNMBP
|
dynamin binding protein
|
|
chr17_-_77824849
|
0.900
|
NM_001893
NM_139062
|
CSNK1D
|
casein kinase 1, delta
|
|
chr16_+_56211410
|
0.899
|
NM_001145770
NM_005682
|
GPR56
|
G protein-coupled receptor 56
|
|
chr1_+_27541086
|
0.898
|
|
SYTL1
|
synaptotagmin-like 1
|
|
chr20_-_22513050
|
0.865
|
NM_021784
|
FOXA2
|
forkhead box A2
|
|
chr18_-_26935883
|
0.858
|
NM_004949
NM_024422
|
DSC2
|
desmocollin 2
|
|
chr14_-_22974707
|
0.800
|
NM_000257
|
MYH7
|
myosin, heavy chain 7, cardiac muscle, beta
|
|
chr16_+_67236730
|
0.798
|
|
CDH3
|
cadherin 3, type 1, P-cadherin (placental)
|
|
chr17_+_45965044
|
0.791
|
NM_017957
|
EPN3
|
epsin 3
|
|
chr17_-_37144408
|
0.787
|
NM_001079870
NM_001079871
NM_177977
|
HAP1
|
huntingtin-associated protein 1
|
|
chr20_-_42457717
|
0.782
|
|
|
|
|
chr16_+_67236699
|
0.781
|
|
CDH3
|
cadherin 3, type 1, P-cadherin (placental)
|
|
chr16_+_56211150
|
0.768
|
|
GPR56
|
G protein-coupled receptor 56
|
|
chr17_+_33761234
|
0.751
|
NM_014598
|
SOCS7
|
suppressor of cytokine signaling 7
|
|
chr1_-_150070849
|
0.750
|
NM_005060
|
RORC
|
RAR-related orphan receptor C
|
|
chr5_+_129268352
|
0.749
|
NM_175856
|
CHSY3
|
chondroitin sulfate synthase 3
|
|
chr14_-_22596522
|
0.744
|
NM_022478
NM_144985
|
CDH24
|
cadherin 24, type 2
|
|
chr6_-_30151526
|
0.720
|
NM_170769
NM_025236
|
RNF39
|
ring finger protein 39
|
|
chr4_-_118226119
|
0.698
|
NM_152402
|
TRAM1L1
|
translocation associated membrane protein 1-like 1
|
|
chr20_-_22514092
|
0.690
|
NM_153675
|
FOXA2
|
forkhead box A2
|
|
chr2_-_219633481
|
0.688
|
NM_002181
|
IHH
|
Indian hedgehog
|
|
chr19_+_40431678
|
0.687
|
|
LSR
|
lipolysis stimulated lipoprotein receptor
|
|
chr16_-_66535923
|
0.686
|
|
SLC12A4
|
solute carrier family 12 (potassium/chloride transporters), member 4
|
|
chr12_+_39372456
|
0.677
|
NM_001843
NM_175038
|
CNTN1
|
contactin 1
|
|
chr17_-_40858775
|
0.671
|
NM_199282
|
ARHGAP27
|
Rho GTPase activating protein 27
|
|
chr3_+_125786195
|
0.664
|
NM_007064
|
KALRN
|
kalirin, RhoGEF kinase
|
|
chr16_+_29373365
|
0.654
|
NM_001014999
NM_001015000
NM_024044
NM_178044
|
SLX1A-SULT1A3
SLX1A
SLX1B
|
SLX1A-SULT1A3 read-through transcript
SLX1 structure-specific endonuclease subunit homolog A (S. cerevisiae)
SLX1 structure-specific endonuclease subunit homolog B (S. cerevisiae)
|
|
chr14_+_23937766
|
0.646
|
NM_025081
|
NYNRIN
|
NYN domain and retroviral integrase containing
|
|
chr9_+_135389698
|
0.641
|
NM_014694
|
ADAMTSL2
|
ADAMTS-like 2
|
|
chr7_+_45894480
|
0.635
|
NM_000596
|
IGFBP1
|
insulin-like growth factor binding protein 1
|
|
chr2_-_42873179
|
0.635
|
NM_012205
|
HAAO
|
3-hydroxyanthranilate 3,4-dioxygenase
|
|
chr2_+_242400702
|
0.633
|
NM_001167599
NM_001167602
NM_080741
|
NEU4
|
sialidase 4
|
|
chr20_-_9767406
|
0.631
|
NM_020341
NM_177990
|
PAK7
|
p21 protein (Cdc42/Rac)-activated kinase 7
|
|
chr11_+_12987455
|
0.623
|
NM_001080521
|
RASSF10
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 10
|
|
chr11_-_70641267
|
0.618
|
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr14_-_24588916
|
0.611
|
NM_014178
|
STXBP6
|
syntaxin binding protein 6 (amisyn)
|
|
chr9_+_115957537
|
0.606
|
|
COL27A1
|
collagen, type XXVII, alpha 1
|
|
chr12_-_52980914
|
0.598
|
|
NFE2
|
nuclear factor (erythroid-derived 2), 45kDa
|
|
chr13_-_33148899
|
0.598
|
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr16_-_4778317
|
0.593
|
NM_001154458
NM_144605
|
SEPT12
|
septin 12
|
|
chr17_+_45405128
|
0.590
|
NM_001934
|
DLX4
|
distal-less homeobox 4
|
|
chr16_-_31054321
|
0.587
|
|
PRSS8
|
protease, serine, 8
|
|
chr17_+_71582486
|
0.584
|
NM_003857
|
GALR2
|
galanin receptor 2
|
|
chr3_-_150858274
|
0.582
|
NM_001168280
NM_015472
|
WWTR1
|
WW domain containing transcription regulator 1
|
|
chr20_+_31334601
|
0.577
|
NM_033197
|
C20orf114
|
chromosome 20 open reading frame 114
|
|
chr1_-_24307514
|
0.565
|
|
MYOM3
|
myomesin family, member 3
|
|
chr12_+_48784057
|
0.565
|
NM_005276
|
GPD1
|
glycerol-3-phosphate dehydrogenase 1 (soluble)
|
|
chr1_-_150079656
|
0.556
|
NM_001136003
|
C2CD4D
|
C2 calcium-dependent domain containing 4D
|
|
chr10_-_105411339
|
0.552
|
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr2_-_165186232
|
0.548
|
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chr16_+_65765256
|
0.543
|
NM_003946
NM_001185057
|
NOL3
|
nucleolar protein 3 (apoptosis repressor with CARD domain)
|
|
chr6_-_31954730
|
0.542
|
|
SLC44A4
|
solute carrier family 44, member 4
|
|
chr11_-_2116775
|
0.538
|
NM_000612
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr7_-_142816019
|
0.538
|
NM_005232
|
EPHA1
|
EPH receptor A1
|
|
chr13_+_92677012
|
0.536
|
NM_005708
|
GPC6
|
glypican 6
|
|
chr12_+_64504835
|
0.534
|
|
HMGA2
|
high mobility group AT-hook 2
|
|
chr20_-_22512893
|
0.534
|
|
FOXA2
|
forkhead box A2
|
|
chr3_+_214668
|
0.533
|
|
|
|
|
chr7_-_20791907
|
0.532
|
|
|
|
|
chr3_-_184316553
|
0.531
|
|
MCCC1
|
methylcrotonoyl-CoA carboxylase 1 (alpha)
|
|
chr16_-_49742008
|
0.529
|
NM_001127892
|
SALL1
|
sal-like 1 (Drosophila)
|
|
chr2_-_165186603
|
0.529
|
NM_004490
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chr1_-_159459999
|
0.528
|
NM_001643
|
APOA2
|
apolipoprotein A-II
|
|
chr5_+_137829077
|
0.526
|
NM_001964
|
EGR1
|
early growth response 1
|
|
chr15_-_81412354
|
0.525
|
NM_004839
NM_199330
NM_199331
NM_199332
|
HOMER2
|
homer homolog 2 (Drosophila)
|
|
chr4_-_15694407
|
0.522
|
|
PROM1
|
prominin 1
|
|
chr1_-_151855413
|
0.519
|
NM_020672
|
S100A14
|
S100 calcium binding protein A14
|
|
chr14_-_22516271
|
0.516
|
NM_198086
|
JUB
|
jub, ajuba homolog (Xenopus laevis)
|
|
chr16_+_2954217
|
0.510
|
NM_024507
NM_172229
|
KREMEN2
|
kringle containing transmembrane protein 2
|
|
chr9_+_676640
|
0.508
|
|
|
|
|
chr12_+_106236281
|
0.506
|
NM_001018072
|
BTBD11
|
BTB (POZ) domain containing 11
|
|
chr14_+_89597860
|
0.506
|
NM_022054
|
KCNK13
|
potassium channel, subfamily K, member 13
|
|
chr17_-_74231266
|
0.503
|
|
CYTH1
|
cytohesin 1
|
|
chr22_+_29122932
|
0.500
|
NM_012429
NM_033382
|
SEC14L2
|
SEC14-like 2 (S. cerevisiae)
|
|
chr12_-_49878216
|
0.495
|
NM_002702
|
POU6F1
|
POU class 6 homeobox 1
|
|
chr6_+_31538934
|
0.495
|
NM_006674
|
HCP5
|
HLA complex P5
|
|
chr1_+_27541049
|
0.494
|
NM_001193308
NM_032872
|
SYTL1
|
synaptotagmin-like 1
|
|
chr12_-_47468985
|
0.494
|
NM_020983
|
ADCY6
|
adenylate cyclase 6
|
|
chr17_-_67628523
|
0.494
|
|
FLJ37644
|
hypothetical LOC400618
|
|
chr20_-_17610704
|
0.493
|
|
RRBP1
|
ribosome binding protein 1 homolog 180kDa (dog)
|
|
chr11_-_116168316
|
0.493
|
NM_001166598
NM_052968
|
APOA5
|
apolipoprotein A-V
|
|
chr6_+_6670117
|
0.491
|
|
|
|
|
chr12_-_193516
|
0.491
|
NM_001122848
|
SLC6A12
|
solute carrier family 6 (neurotransmitter transporter, betaine/GABA), member 12
|
|
chr18_-_22383066
|
0.486
|
NM_001136205
|
KCTD1
|
potassium channel tetramerisation domain containing 1
|
|
chr16_-_31054563
|
0.486
|
NM_002773
|
PRSS8
|
protease, serine, 8
|
|
chr19_+_45973139
|
0.484
|
NM_006533
|
MIA
|
melanoma inhibitory activity
|
|
chr11_+_45900731
|
0.482
|
|
GYLTL1B
|
glycosyltransferase-like 1B
|
|
chr1_-_224143389
|
0.481
|
NM_020997
|
LEFTY1
|
left-right determination factor 1
|
|
chr14_+_70178126
|
0.481
|
NM_015351
|
TTC9
|
tetratricopeptide repeat domain 9
|
|
chr16_+_12902955
|
0.480
|
NM_001145204
NM_001145205
|
SHISA9
|
shisa homolog 9 (Xenopus laevis)
|
|
chr9_-_106705867
|
0.478
|
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr8_-_144726064
|
0.477
|
NM_001100878
|
C8orf73
|
chromosome 8 open reading frame 73
|
|
chr12_+_48652886
|
0.476
|
NM_001652
|
AQP6
|
aquaporin 6, kidney specific
|
|
chr5_+_79021401
|
0.468
|
NM_153610
|
CMYA5
|
cardiomyopathy associated 5
|
|
chr19_-_6671585
|
0.463
|
NM_000064
|
C3
|
complement component 3
|
|
chrX_-_107866262
|
0.458
|
NM_003604
|
IRS4
|
insulin receptor substrate 4
|
|
chr16_-_49742651
|
0.456
|
NM_002968
|
SALL1
|
sal-like 1 (Drosophila)
|
|
chr14_+_36196523
|
0.456
|
NM_006194
|
PAX9
|
paired box 9
|
|
chr17_+_69939806
|
0.453
|
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C
|
|
chrX_-_20044670
|
0.451
|
NM_001168467
|
MAP7D2
|
MAP7 domain containing 2
|
|
chr2_+_70981223
|
0.450
|
NM_012476
|
VAX2
|
ventral anterior homeobox 2
|
|
chr10_-_71753684
|
0.449
|
|
LRRC20
|
leucine rich repeat containing 20
|
|
chr6_-_24466258
|
0.448
|
NM_016356
|
DCDC2
|
doublecortin domain containing 2
|
|
chr20_+_20296744
|
0.447
|
NM_002196
|
INSM1
|
insulinoma-associated 1
|
|
chr6_+_43036442
|
0.446
|
NM_018960
|
GNMT
|
glycine N-methyltransferase
|
|
chr12_+_439803
|
0.443
|
NM_173593
|
B4GALNT3
|
beta-1,4-N-acetyl-galactosaminyl transferase 3
|
|
chr10_+_123862430
|
0.443
|
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr22_-_44751671
|
0.442
|
NM_058238
|
WNT7B
|
wingless-type MMTV integration site family, member 7B
|
|
chr6_+_32090516
|
0.438
|
NM_007293
NM_001002029
|
C4A
C4B
|
complement component 4A (Rodgers blood group)
complement component 4B (Chido blood group)
|
|
chr1_-_21868380
|
0.437
|
NM_001145657
NM_002885
|
RAP1GAP
|
RAP1 GTPase activating protein
|
|
chr19_+_6415259
|
0.437
|
NM_139161
NM_174881
|
CRB3
|
crumbs homolog 3 (Drosophila)
|
|
chr2_-_165186064
|
0.437
|
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chr15_-_71448194
|
0.434
|
|
HCN4
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 4
|
|
chr15_-_72804763
|
0.431
|
NM_000499
|
CYP1A1
|
cytochrome P450, family 1, subfamily A, polypeptide 1
|
|
chr3_+_152287365
|
0.430
|
NM_053002
|
MED12L
|
mediator complex subunit 12-like
|
|
chr16_-_51138306
|
0.429
|
NM_001080430
|
TOX3
|
TOX high mobility group box family member 3
|
|
chr7_+_100156358
|
0.429
|
NM_000799
|
EPO
|
erythropoietin
|
|
chr1_+_15955719
|
0.428
|
|
FBLIM1
|
filamin binding LIM protein 1
|
|
chr8_-_144887876
|
0.428
|
NM_198488
|
FAM83H
|
family with sequence similarity 83, member H
|
|
chr6_+_41714171
|
0.427
|
NM_005586
|
MDFI
|
MyoD family inhibitor
|
|
chr15_-_68933481
|
0.426
|
NM_018357
NM_197958
|
LARP6
|
La ribonucleoprotein domain family, member 6
|
|
chr1_+_199127242
|
0.422
|
NM_018265
|
C1orf106
|
chromosome 1 open reading frame 106
|
|
chr16_-_28425632
|
0.421
|
NM_145659
|
IL27
|
interleukin 27
|
|
chr15_-_71448657
|
0.420
|
NM_005477
|
HCN4
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 4
|
|
chr7_-_96492064
|
0.419
|
NM_005221
|
DLX5
|
distal-less homeobox 5
|
|
chr2_-_139254273
|
0.418
|
NM_007226
|
NXPH2
|
neurexophilin 2
|
|
chr2_+_70981233
|
0.418
|
|
VAX2
|
ventral anterior homeobox 2
|
|
chr4_-_120768563
|
0.416
|
NM_033437
|
PDE5A
|
phosphodiesterase 5A, cGMP-specific
|
|
chr9_-_106705780
|
0.415
|
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr17_+_35425139
|
0.411
|
NM_000759
NM_001178147
NM_172219
NM_172220
|
CSF3
|
colony stimulating factor 3 (granulocyte)
|
|
chrX_-_106033202
|
0.409
|
NM_001171706
NM_138382
|
RIPPLY1
|
ripply1 homolog (zebrafish)
|
|
chr17_-_36469869
|
0.408
|
NM_001165252
|
LOC730755
|
keratin associated protein 2-4-like
|
|
chr10_-_118419459
|
0.408
|
NM_144661
|
C10orf82
|
chromosome 10 open reading frame 82
|
|
chr9_-_85342868
|
0.407
|
NM_174938
|
FRMD3
|
FERM domain containing 3
|
|
chr11_-_8788765
|
0.402
|
NM_213618
|
ST5
|
suppression of tumorigenicity 5
|
|
chr6_+_30082331
|
0.402
|
|
HLA-J
|
major histocompatibility complex, class I, J (pseudogene)
|
|
chr3_+_10832867
|
0.401
|
NM_014229
|
SLC6A11
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 11
|
|
chr3_-_9265592
|
0.401
|
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating protein 3
|
|
chr19_+_49972965
|
0.400
|
NM_001130852
NM_012116
|
CBLC
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence c
|
|
chr11_+_62893836
|
0.400
|
NM_080866
|
SLC22A9
|
solute carrier family 22 (organic anion transporter), member 9
|
|
chr4_-_1156362
|
0.396
|
|
SPON2
|
spondin 2, extracellular matrix protein
|
|
chr19_+_40223249
|
0.395
|
NM_002151
NM_182983
|
HPN
|
hepsin
|
|
chr1_+_57093030
|
0.394
|
NM_000562
|
C8A
|
complement component 8, alpha polypeptide
|
|
chr20_-_61933012
|
0.393
|
|
ZBTB46
|
zinc finger and BTB domain containing 46
|
|
chr14_-_76677711
|
0.390
|
|
ZDHHC22
|
zinc finger, DHHC-type containing 22
|
|
chr20_+_3399615
|
0.385
|
NM_139321
NM_139322
|
ATRN
|
attractin
|
|
chr6_-_18230829
|
0.384
|
NM_198586
|
NHLRC1
|
NHL repeat containing 1
|
|
chr20_-_17610828
|
0.384
|
NM_001042576
NM_004587
|
RRBP1
|
ribosome binding protein 1 homolog 180kDa (dog)
|
|
chr1_+_24755152
|
0.383
|
NM_001010980
|
C1orf130
|
chromosome 1 open reading frame 130
|
|
chr14_-_23781607
|
0.381
|
NM_001099274
NM_012461
|
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2
|
|
chr5_-_134899537
|
0.379
|
NM_006161
|
NEUROG1
|
neurogenin 1
|
|
chr2_+_219532590
|
0.379
|
NM_003936
|
CDK5R2
|
cyclin-dependent kinase 5, regulatory subunit 2 (p39)
|
|
chr9_-_4289574
|
0.379
|
|
GLIS3
|
GLIS family zinc finger 3
|
|
chr3_-_187562470
|
0.378
|
NM_001080744
NM_001080745
NM_001346
|
DGKG
|
diacylglycerol kinase, gamma 90kDa
|
|
chr14_-_20636337
|
0.378
|
|
ZNF219
|
zinc finger protein 219
|
|
chr7_-_99411573
|
0.377
|
NM_001185
|
AZGP1
|
alpha-2-glycoprotein 1, zinc-binding
|
|
chrX_-_132376660
|
0.376
|
NM_001448
|
GPC4
|
glypican 4
|
|
chr21_-_36774257
|
0.376
|
NM_001146079
NM_144492
|
CLDN14
|
claudin 14
|
|
chr5_+_5193442
|
0.376
|
NM_139056
|
ADAMTS16
|
ADAM metallopeptidase with thrombospondin type 1 motif, 16
|
|
chrX_-_132947290
|
0.375
|
NM_001164617
NM_001164618
NM_001164619
NM_004484
|
GPC3
|
glypican 3
|
|
chr2_-_86889029
|
0.374
|
NM_001145873
|
CD8A
|
CD8a molecule
|
|
chr9_+_70926009
|
0.371
|
NM_001170414
|
TJP2
|
tight junction protein 2 (zona occludens 2)
|
|
chr12_-_24606616
|
0.366
|
NM_152989
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr19_+_39664706
|
0.366
|
NM_001080436
|
WTIP
|
Wilms tumor 1 interacting protein
|
|
chr11_-_124127343
|
0.364
|
|
VSIG2
|
V-set and immunoglobulin domain containing 2
|
|
chr1_+_46418786
|
0.364
|
|
TSPAN1
|
tetraspanin 1
|
|
chrX_-_132947141
|
0.363
|
|
GPC3
|
glypican 3
|
|
chr14_-_23686269
|
0.363
|
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta)
|
|
chr11_-_124127295
|
0.363
|
NM_014312
|
VSIG2
|
V-set and immunoglobulin domain containing 2
|
|
chr9_+_111582317
|
0.360
|
NM_053016
NM_007203
NM_147150
|
PALM2
PALM2-AKAP2
|
paralemmin 2
PALM2-AKAP2 readthrough
|
|
chr17_-_35017650
|
0.360
|
|
NEUROD2
|
neurogenic differentiation 2
|
|
chr15_+_58083798
|
0.360
|
|
FOXB1
|
forkhead box B1
|
|
chr12_+_47658502
|
0.359
|
NM_005430
|
WNT1
|
wingless-type MMTV integration site family, member 1
|
|
chr16_+_54247042
|
0.358
|
NM_001172501
|
SLC6A2
|
solute carrier family 6 (neurotransmitter transporter, noradrenalin), member 2
|
|
chrX_-_54401161
|
0.357
|
NM_001002838
NM_020922
|
WNK3
|
WNK lysine deficient protein kinase 3
|
|
chr14_+_23527866
|
0.356
|
NM_198083
|
DHRS4
DHRS4L2
|
dehydrogenase/reductase (SDR family) member 4
dehydrogenase/reductase (SDR family) member 4 like 2
|
|
chr19_-_63715135
|
0.355
|
NM_012254
|
SLC27A5
|
solute carrier family 27 (fatty acid transporter), member 5
|
|
chr2_-_218843109
|
0.354
|
NM_001087
|
AAMP
|
angio-associated, migratory cell protein
|
|
chr20_+_1824006
|
0.353
|
|
SIRPA
|
signal-regulatory protein alpha
|
|
chr3_+_85090721
|
0.351
|
NM_001167674
NM_001167675
|
CADM2
|
cell adhesion molecule 2
|
|
chr22_+_18330049
|
0.351
|
NM_007310
|
COMT
|
catechol-O-methyltransferase
|
|
chr17_-_36928604
|
0.350
|
NM_002275
|
KRT15
|
keratin 15
|
|
chr18_-_23019176
|
0.350
|
NM_031422
|
CHST9
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 9
|
|
chr20_-_581844
|
0.350
|
NM_080725
|
SRXN1
|
sulfiredoxin 1
|
|
chr3_+_187840842
|
0.349
|
NM_014375
|
FETUB
|
fetuin B
|
|
chr6_+_160689355
|
0.345
|
NM_021977
|
SLC22A3
|
solute carrier family 22 (extraneuronal monoamine transporter), member 3
|
|
chr2_+_219779737
|
0.344
|
NM_138802
|
ZFAND2B
|
zinc finger, AN1-type domain 2B
|
|
chr2_-_26058941
|
0.343
|
NM_002254
|
KIF3C
|
kinesin family member 3C
|
|
chr17_+_45993338
|
0.343
|
NM_018896
NM_198376
NM_198377
NM_198378
NM_198379
NM_198380
NM_198382
NM_198383
NM_198384
NM_198385
NM_198386
NM_198387
NM_198388
NM_198396
NM_198397
|
CACNA1G
|
calcium channel, voltage-dependent, T type, alpha 1G subunit
|
|
chr16_-_14695930
|
0.343
|
NM_003561
|
PLA2G10
|
phospholipase A2, group X
|
|
chr11_-_124127244
|
0.340
|
|
VSIG2
|
V-set and immunoglobulin domain containing 2
|