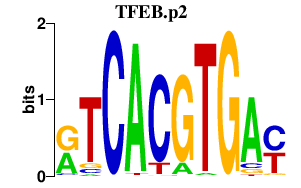
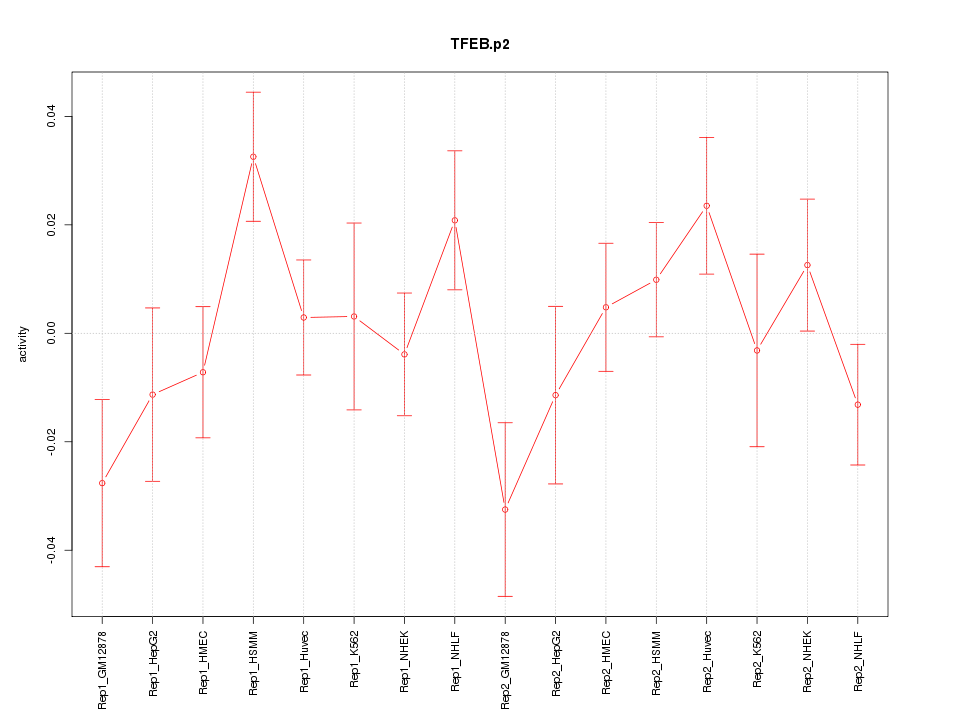
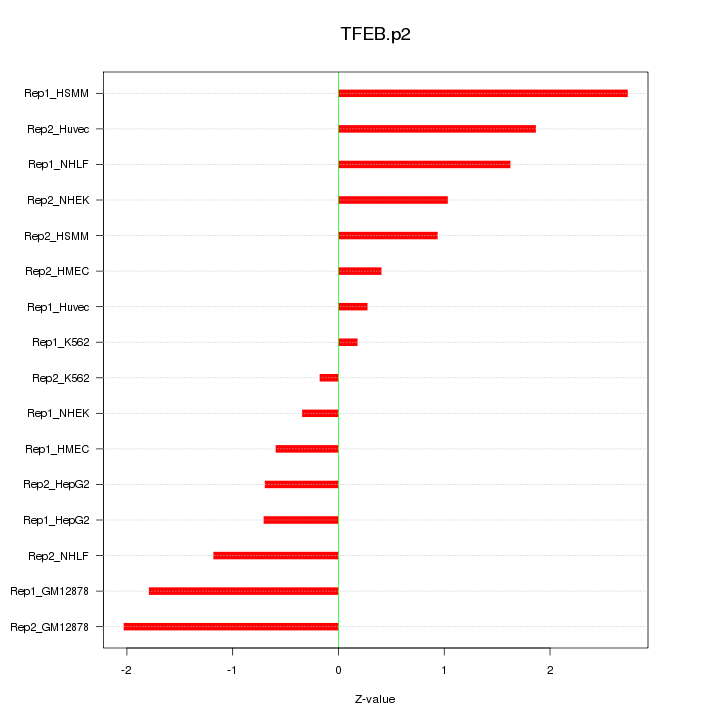
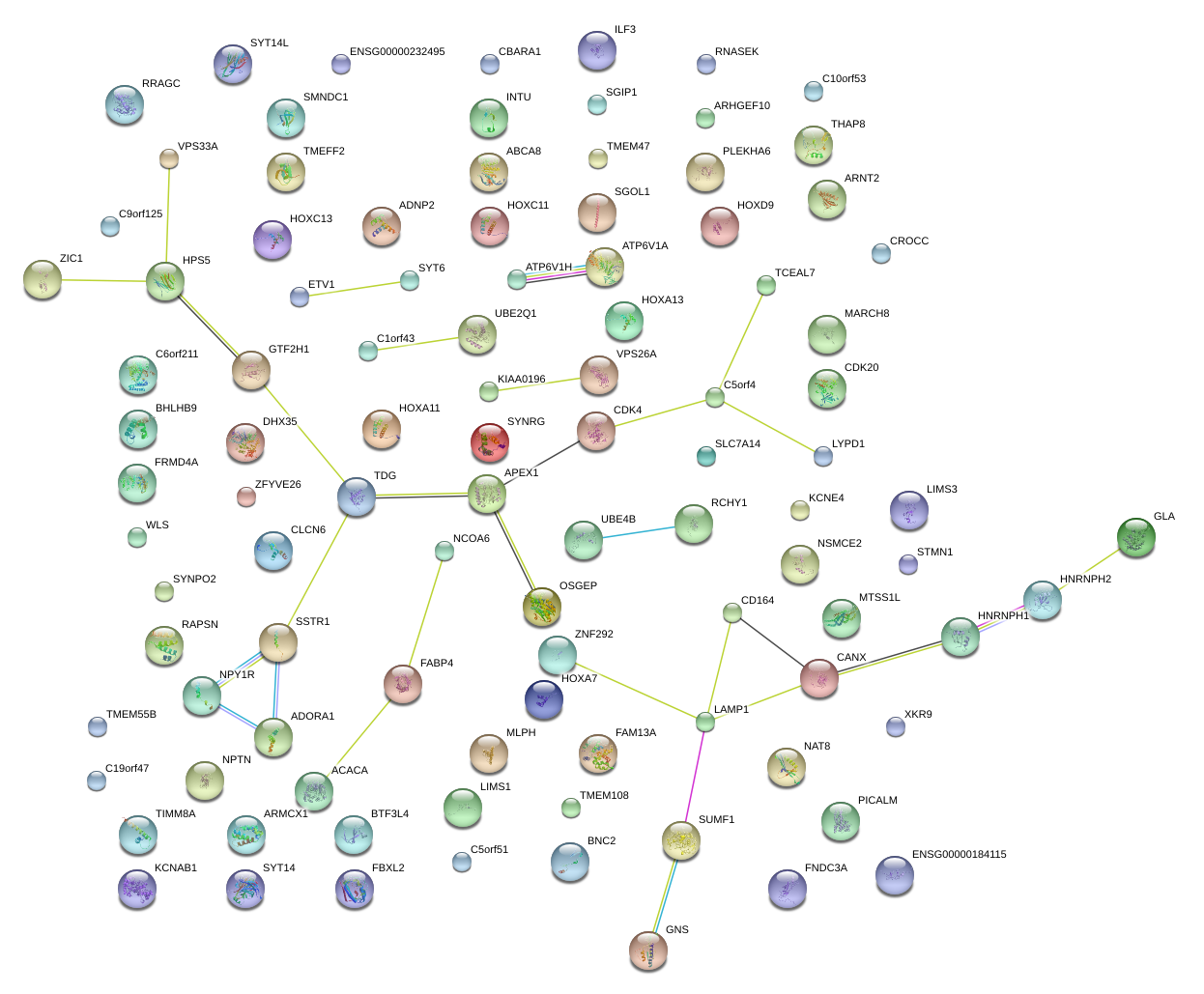
|
chr6_+_87921980
|
3.249
|
NM_015021
|
ZNF292
|
zinc finger protein 292
|
|
chr10_-_14090459
|
2.739
|
|
FRMD4A
|
FERM domain containing 4A
|
|
chr7_-_27162688
|
2.525
|
NM_006896
|
HOXA7
|
homeobox A7
|
|
chr1_-_152459506
|
2.331
|
|
C1orf43
|
chromosome 1 open reading frame 43
|
|
chr1_-_202595666
|
2.300
|
NM_014935
|
PLEKHA6
|
pleckstrin homology domain containing, family A member 6
|
|
chr1_-_68470585
|
2.253
|
NM_001002292
NM_001193334
NM_024911
|
WLS
|
wntless homolog (Drosophila)
|
|
chrX_+_101862297
|
2.173
|
NM_001142524
NM_001142525
NM_001142526
NM_001142527
NM_001142528
NM_030639
|
BHLHB9
|
basic helix-loop-helix domain containing, class B, 9
|
|
chr7_+_27191534
|
2.031
|
|
HOXA11-AS1
|
HOXA11 antisense RNA 1 (non-protein coding)
|
|
chr1_-_152459667
|
1.984
|
|
C1orf43
|
chromosome 1 open reading frame 43
|
|
chr11_-_18300246
|
1.915
|
NM_007216
NM_181507
NM_181508
|
HPS5
|
Hermansky-Pudlak syndrome 5
|
|
chr19_+_10626106
|
1.885
|
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa
|
|
chr2_+_223625105
|
1.828
|
NM_080671
|
KCNE4
|
potassium voltage-gated channel, Isk-related family, member 4
|
|
chr1_+_201364050
|
1.800
|
|
ADORA1
|
adenosine A1 receptor
|
|
chr3_+_134239860
|
1.799
|
NM_001136469
NM_023943
|
TMEM108
|
transmembrane protein 108
|
|
chr2_-_133144094
|
1.775
|
|
LYPD1
|
LY6/PLAUR domain containing 1
|
|
chr10_+_70553993
|
1.712
|
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr3_+_157491469
|
1.690
|
NM_172159
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1
|
|
chr3_+_134239906
|
1.628
|
|
TMEM108
|
transmembrane protein 108
|
|
chr12_+_52653176
|
1.627
|
NM_014212
|
HOXC11
|
homeobox C11
|
|
chr8_+_126173264
|
1.618
|
NM_173685
|
NSMCE2
|
non-SMC element 2, MMS21 homolog (S. cerevisiae)
|
|
chr1_-_152797820
|
1.611
|
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|
|
chrX_-_34585287
|
1.610
|
NM_031442
|
TMEM47
|
transmembrane protein 47
|
|
chr3_-_20202622
|
1.583
|
NM_001012409
NM_001012410
NM_001012411
NM_001012412
NM_001012413
NM_138484
|
SGOL1
|
shugoshin-like 1 (S. pombe)
|
|
chrX_+_102471769
|
1.580
|
NM_152278
|
TCEAL7
|
transcription elongation factor A (SII)-like 7
|
|
chr8_-_126173055
|
1.575
|
|
KIAA0196
|
KIAA0196
|
|
chr9_-_89779413
|
1.575
|
NM_001039803
NM_001170639
NM_001170640
NM_012119
NM_178432
|
CDK20
|
cyclin-dependent kinase 20
|
|
chr10_+_70553936
|
1.565
|
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr9_-_16860719
|
1.565
|
NM_017637
|
BNC2
|
basonuclin 2
|
|
chr2_+_238060602
|
1.559
|
NM_001042467
NM_024101
|
MLPH
|
melanophilin
|
|
chr4_+_120167721
|
1.557
|
|
SYNPO2
|
synaptopodin 2
|
|
chr11_-_85457469
|
1.544
|
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr7_-_27191287
|
1.534
|
NM_005523
|
HOXA11
|
homeobox A11
|
|
chr8_-_126173108
|
1.529
|
|
KIAA0196
|
KIAA0196
|
|
chr3_-_171786468
|
1.512
|
NM_020949
|
SLC7A14
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 14
|
|
chr4_+_128773529
|
1.507
|
NM_015693
|
INTU
|
inturned planar cell polarity effector homolog (Drosophila)
|
|
chr14_-_19993032
|
1.481
|
NM_017807
|
OSGEP
|
O-sialoglycoprotein endopeptidase
|
|
chr14_+_37746937
|
1.436
|
NM_001049
|
SSTR1
|
somatostatin receptor 1
|
|
chr3_+_148609841
|
1.404
|
NM_003412
|
ZIC1
|
Zic family member 1 (odd-paired homolog, Drosophila)
|
|
chr1_+_66772393
|
1.380
|
NM_032291
|
SGIP1
|
SH3-domain GRB2-like (endophilin) interacting protein 1
|
|
chr11_-_47427263
|
1.371
|
NM_005055
NM_032645
|
RAPSN
|
receptor-associated protein of the synapse
|
|
chr8_-_126173186
|
1.351
|
|
KIAA0196
|
KIAA0196
|
|
chr4_-_90197140
|
1.341
|
NM_014883
|
FAM13A
|
family with sequence similarity 13, member A
|
|
chr9_-_16860664
|
1.336
|
|
BNC2
|
basonuclin 2
|
|
chr1_+_17121012
|
1.329
|
NM_014675
|
CROCC
|
ciliary rootlet coiled-coil, rootletin
|
|
chr13_+_112999590
|
1.326
|
|
LAMP1
|
lysosomal-associated membrane protein 1
|
|
chr5_+_41940216
|
1.316
|
NM_175921
|
C5orf51
|
chromosome 5 open reading frame 51
|
|
chr16_-_69277355
|
1.302
|
NM_138383
|
MTSS1L
|
metastasis suppressor 1-like
|
|
chr11_-_71491916
|
1.292
|
|
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1
|
|
chr8_-_126173203
|
1.289
|
|
KIAA0196
|
KIAA0196
|
|
chr2_+_108571177
|
1.282
|
NM_004987
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chrX_-_114703273
|
1.281
|
|
|
|
|
chr9_-_103289154
|
1.278
|
NM_032342
|
C9orf125
|
chromosome 9 open reading frame 125
|
|
chr8_-_126173189
|
1.277
|
|
KIAA0196
|
KIAA0196
|
|
chr3_-_4483939
|
1.261
|
|
SUMF1
|
sulfatase modifying factor 1
|
|
chr11_+_18300370
|
1.255
|
NM_001142307
NM_005316
|
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa
|
|
chr10_+_70553983
|
1.250
|
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr4_-_164473107
|
1.232
|
NM_000909
|
NPY1R
|
neuropeptide Y receptor Y1
|
|
chr1_-_152797760
|
1.192
|
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|
|
chr15_-_71712711
|
1.190
|
NM_001161363
NM_001161364
NM_012428
NM_017455
|
NPTN
|
neuroplastin
|
|
chr2_+_176695658
|
1.189
|
NM_014213
|
HOXD9
|
homeobox D9
|
|
chr14_+_19993181
|
1.185
|
|
APEX1
|
APEX nuclease (multifunctional DNA repair enzyme) 1
|
|
chr8_+_1759502
|
1.184
|
NM_014629
|
ARHGEF10
|
Rho guanine nucleotide exchange factor (GEF) 10
|
|
chr4_+_120167770
|
1.181
|
|
SYNPO2
|
synaptopodin 2
|
|
chr19_-_41236959
|
1.176
|
|
THAP8
|
THAP domain containing 8
|
|
chr17_-_33043514
|
1.175
|
NM_001163544
NM_001163545
NM_001163546
NM_001163547
NM_007247
NM_080550
NM_198882
|
SYNRG
|
synergin, gamma
|
|
chr19_+_10625987
|
1.169
|
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa
|
|
chr11_+_18300689
|
1.164
|
|
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa
|
|
chr3_+_114948550
|
1.154
|
NM_001690
|
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A
|
|
chr1_+_208178160
|
1.154
|
NM_001146261
NM_001146262
NM_001146264
NM_153262
|
SYT14
|
synaptotagmin XIV
|
|
chr1_-_114497994
|
1.142
|
NM_205848
|
SYT6
|
synaptotagmin VI
|
|
chr6_+_151815298
|
1.138
|
|
C6orf211
|
chromosome 6 open reading frame 211
|
|
chr6_-_169392771
|
1.133
|
|
|
|
|
chr17_+_6856697
|
1.128
|
|
RNASEK-C17ORF49
|
RNASEK-C17orf49 read-through transcript
|
|
chr12_-_121316907
|
1.114
|
|
VPS33A
|
vacuolar protein sorting 33 homolog A (S. cerevisiae)
|
|
chr6_+_151815114
|
1.098
|
NM_024573
|
C6orf211
|
chromosome 6 open reading frame 211
|
|
chr13_+_48582389
|
1.098
|
NM_014923
|
FNDC3A
|
fibronectin type III domain containing 3A
|
|
chr12_-_56432377
|
1.095
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr17_+_6856676
|
1.095
|
|
RNASEK-C17ORF49
RNASEK
|
RNASEK-C17orf49 read-through transcript
ribonuclease, RNase K
|
|
chr8_-_54918099
|
1.089
|
NM_213620
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H
|
|
chr11_-_85457736
|
1.078
|
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chrX_-_100549444
|
1.074
|
|
GLA
|
galactosidase, alpha
|
|
chr9_-_89779180
|
1.073
|
|
CDK20
|
cyclin-dependent kinase 20
|
|
chr2_+_108571209
|
1.057
|
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr8_-_126173227
|
1.054
|
NM_014846
|
KIAA0196
|
KIAA0196
|
|
chr7_-_13997389
|
1.052
|
NM_001163149
|
ETV1
|
ets variant 1
|
|
chr10_+_70553945
|
1.043
|
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr6_-_109810246
|
1.034
|
|
CD164
|
CD164 molecule, sialomucin
|
|
chrX_+_100692071
|
1.033
|
NM_016608
|
ARMCX1
|
armadillo repeat containing, X-linked 1
|
|
chr17_-_32730804
|
1.029
|
NM_198837
NM_198838
|
ACACA
|
acetyl-CoA carboxylase alpha
|
|
chr2_-_192767844
|
1.022
|
NM_016192
|
TMEFF2
|
transmembrane protein with EGF-like and two follistatin-like domains 2
|
|
chr10_+_70553949
|
1.019
|
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr12_-_56432375
|
1.017
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr14_+_19993204
|
1.017
|
|
APEX1
|
APEX nuclease (multifunctional DNA repair enzyme) 1
|
|
chr1_-_26105230
|
1.014
|
NM_203401
|
STMN1
|
stathmin 1
|
|
chr6_-_109810296
|
1.013
|
|
CD164
|
CD164 molecule, sialomucin
|
|
chr3_-_4483928
|
1.003
|
|
SUMF1
|
sulfatase modifying factor 1
|
|
chr1_-_152459697
|
0.999
|
NM_001098616
NM_015449
NM_138740
|
C1orf43
|
chromosome 1 open reading frame 43
|
|
chr6_-_109810396
|
0.990
|
NM_001142401
NM_001142402
NM_001142403
NM_001142404
NM_006016
|
CD164
|
CD164 molecule, sialomucin
|
|
chr6_-_109810310
|
0.989
|
|
CD164
|
CD164 molecule, sialomucin
|
|
chr1_-_39097904
|
0.983
|
NM_022157
|
RRAGC
|
Ras-related GTP binding C
|
|
chr10_-_112054679
|
0.982
|
NM_005871
|
SMNDC1
|
survival motor neuron domain containing 1
|
|
chr11_-_85457753
|
0.973
|
NM_001008660
NM_007166
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr10_-_112054497
|
0.972
|
|
SMNDC1
|
survival motor neuron domain containing 1
|
|
chr3_-_4483952
|
0.972
|
|
SUMF1
|
sulfatase modifying factor 1
|
|
chr11_-_71491942
|
0.967
|
|
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1
|
|
chr6_-_109810354
|
0.966
|
|
CD164
|
CD164 molecule, sialomucin
|
|
chr3_-_4483955
|
0.963
|
NM_001164674
NM_001164675
NM_182760
|
SUMF1
|
sulfatase modifying factor 1
|
|
chr11_-_71491983
|
0.958
|
|
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1
|
|
chr12_+_52618815
|
0.956
|
NM_017410
|
HOXC13
|
homeobox C13
|
|
chr14_-_67353046
|
0.952
|
NM_015346
|
ZFYVE26
|
zinc finger, FYVE domain containing 26
|
|
chr1_-_152797707
|
0.946
|
NM_017582
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|
|
chr14_-_19999324
|
0.929
|
|
TMEM55B
|
transmembrane protein 55B
|
|
chr10_+_70553862
|
0.920
|
NM_001035260
NM_004896
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr10_-_45410197
|
0.916
|
NM_001002265
NM_145021
|
MARCH8
|
membrane-associated ring finger (C3HC4) 8
|
|
chr12_+_102883687
|
0.915
|
NM_003211
|
TDG
|
thymine-DNA glycosylase
|
|
chr8_-_82557956
|
0.910
|
NM_001442
|
FABP4
|
fatty acid binding protein 4, adipocyte
|
|
chr17_-_64462976
|
0.903
|
NM_007168
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8
|
|
chr8_+_71744153
|
0.902
|
NM_001011720
|
XKR9
|
XK, Kell blood group complex subunit-related family, member 9
|
|
chr1_+_68070573
|
0.895
|
|
|
|
|
chr1_+_10015736
|
0.894
|
|
UBE4B
|
ubiquitination factor E4B (UFD2 homolog, yeast)
|
|
chr17_+_6856521
|
0.893
|
NM_001004333
|
RNASEK-C17ORF49
RNASEK
|
RNASEK-C17orf49 read-through transcript
ribonuclease, RNase K
|
|
chrX_+_100549885
|
0.889
|
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H')
|
|
chr5_-_154210405
|
0.887
|
NM_032385
|
C5orf4
|
chromosome 5 open reading frame 4
|
|
chr1_+_52294554
|
0.886
|
|
BTF3L4
|
basic transcription factor 3-like 4
|
|
chr12_-_121316997
|
0.883
|
NM_022916
|
VPS33A
|
vacuolar protein sorting 33 homolog A (S. cerevisiae)
|
|
chr5_-_154210350
|
0.880
|
|
C5orf4
|
chromosome 5 open reading frame 4
|
|
chr4_-_76658606
|
0.880
|
NM_001008925
NM_001009922
NM_015436
|
RCHY1
|
ring finger and CHY zinc finger domain containing 1
|
|
chr5_+_179058615
|
0.878
|
|
CANX
|
calnexin
|
|
chr1_+_11788889
|
0.877
|
|
CLCN6
|
chloride channel 6
|
|
chr5_-_154210341
|
0.870
|
|
C5orf4
|
chromosome 5 open reading frame 4
|
|
chr18_+_75968133
|
0.867
|
|
ADNP2
|
ADNP homeobox 2
|
|
chr10_-_120504267
|
0.858
|
|
|
|
|
chrX_-_100490329
|
0.853
|
NM_001145951
|
TIMM8A
|
translocase of inner mitochondrial membrane 8 homolog A (yeast)
|
|
chr10_-_74055824
|
0.844
|
|
CBARA1
|
calcium binding atopy-related autoantigen 1
|
|
chr3_+_33293915
|
0.843
|
NM_012157
NM_001171713
|
FBXL2
|
F-box and leucine-rich repeat protein 2
|
|
chr10_+_50557689
|
0.838
|
NM_001042427
NM_182554
|
C10orf53
|
chromosome 10 open reading frame 53
|
|
chr12_-_63439449
|
0.837
|
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase
|
|
chr19_-_45546123
|
0.826
|
NM_178830
|
C19orf47
|
chromosome 19 open reading frame 47
|
|
chr20_+_37024383
|
0.824
|
NM_001190809
NM_021931
|
DHX35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35
|
|
chr1_+_52294563
|
0.823
|
|
BTF3L4
|
basic transcription factor 3-like 4
|
|
chr12_-_56432385
|
0.822
|
NM_000075
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr3_+_114948600
|
0.821
|
|
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A
|
|
chr11_-_71492019
|
0.816
|
NM_017907
|
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1
|
|
chr1_+_148521852
|
0.816
|
NM_144697
|
C1orf51
|
chromosome 1 open reading frame 51
|
|
chr5_+_65476418
|
0.815
|
NM_139168
|
SREK1
|
splicing regulatory glutamine/lysine-rich protein 1
|
|
chr1_-_201162991
|
0.813
|
NM_021633
|
KLHL12
|
kelch-like 12 (Drosophila)
|
|
chr1_-_239587090
|
0.812
|
NM_002924
|
RGS7
|
regulator of G-protein signaling 7
|
|
chr11_+_125586828
|
0.801
|
NM_024556
|
FAM118B
|
family with sequence similarity 118, member B
|
|
chr17_-_43977273
|
0.800
|
NM_002145
|
HOXB2
|
homeobox B2
|
|
chr12_+_104025667
|
0.800
|
|
KIAA1033
|
KIAA1033
|
|
chr4_-_23500706
|
0.799
|
NM_013261
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha
|
|
chr6_+_127629677
|
0.799
|
NM_030963
|
RNF146
|
ring finger protein 146
|
|
chr1_-_152797767
|
0.798
|
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|
|
chr3_+_135687641
|
0.795
|
NM_025180
|
CEP63
|
centrosomal protein 63kDa
|
|
chrX_+_55760774
|
0.792
|
NM_006064
NM_016656
|
RRAGB
|
Ras-related GTP binding B
|
|
chr12_-_63439415
|
0.791
|
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase
|
|
chr3_+_135687553
|
0.784
|
NM_001042400
|
CEP63
|
centrosomal protein 63kDa
|
|
chr6_-_151814861
|
0.774
|
NM_017909
|
RMND1
|
required for meiotic nuclear division 1 homolog (S. cerevisiae)
|
|
chr9_+_94127561
|
0.772
|
NM_001012267
|
CENPP
|
centromere protein P
|
|
chr3_-_158700036
|
0.771
|
NM_024621
|
VEPH1
|
ventricular zone expressed PH domain homolog 1 (zebrafish)
|
|
chr7_-_13997574
|
0.771
|
NM_001163148
|
ETV1
|
ets variant 1
|
|
chr12_-_63439456
|
0.770
|
NM_002076
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase
|
|
chr7_+_32501577
|
0.767
|
|
AVL9
|
AVL9 homolog (S. cerevisiase)
|
|
chr7_-_44496703
|
0.767
|
NM_015332
|
NUDCD3
|
NudC domain containing 3
|
|
chr20_+_56318415
|
0.765
|
|
RAB22A
|
RAB22A, member RAS oncogene family
|
|
chr3_-_198509276
|
0.757
|
|
DLG1
|
discs, large homolog 1 (Drosophila)
|
|
chr14_+_19993129
|
0.756
|
NM_001641
NM_080648
NM_080649
|
APEX1
|
APEX nuclease (multifunctional DNA repair enzyme) 1
|
|
chr7_+_32501665
|
0.756
|
NM_015060
|
AVL9
|
AVL9 homolog (S. cerevisiase)
|
|
chr9_-_34038887
|
0.754
|
NM_018449
|
UBAP2
|
ubiquitin associated protein 2
|
|
chr20_+_56318245
|
0.753
|
|
RAB22A
|
RAB22A, member RAS oncogene family
|
|
chr1_-_11788666
|
0.751
|
NM_005957
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H)
|
|
chr3_+_114948591
|
0.749
|
|
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A
|
|
chr14_-_92869113
|
0.745
|
|
BTBD7
|
BTB (POZ) domain containing 7
|
|
chr12_+_12769985
|
0.738
|
NM_001130415
|
APOLD1
|
apolipoprotein L domain containing 1
|
|
chr6_-_33822659
|
0.737
|
NM_001142883
NM_054111
|
IP6K3
|
inositol hexakisphosphate kinase 3
|
|
chr12_+_104025686
|
0.736
|
|
|
|
|
chr12_+_104025675
|
0.735
|
|
KIAA1033
|
KIAA1033
|
|
chr4_+_76658677
|
0.729
|
NM_144721
|
THAP6
|
THAP domain containing 6
|
|
chr8_+_17824772
|
0.729
|
|
PCM1
|
pericentriolar material 1
|
|
chr19_-_45546250
|
0.727
|
|
C19orf47
|
chromosome 19 open reading frame 47
|
|
chr3_-_113763049
|
0.726
|
NM_022488
|
ATG3
|
ATG3 autophagy related 3 homolog (S. cerevisiae)
|
|
chr8_+_98725604
|
0.722
|
|
MTDH
|
metadherin
|
|
chrX_-_100549561
|
0.722
|
NM_000169
|
GLA
|
galactosidase, alpha
|
|
chr8_+_109525050
|
0.721
|
|
TTC35
|
tetratricopeptide repeat domain 35
|
|
chr14_-_91642641
|
0.721
|
|
ATXN3
ATXN8
|
ataxin 3
ataxin 8
|
|
chr12_-_63439322
|
0.720
|
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase
|
|
chr9_-_37894098
|
0.717
|
|
MCART1
|
mitochondrial carrier triple repeat 1
|
|
chr1_-_77920914
|
0.711
|
NM_015534
|
ZZZ3
|
zinc finger, ZZ-type containing 3
|
|
chr12_-_119826073
|
0.711
|
|
SPPL3
|
signal peptide peptidase 3
|
|
chr2_+_176689737
|
0.710
|
NM_002148
|
HOXD10
|
homeobox D10
|
|
chr12_-_9913199
|
0.710
|
|
CLEC2B
|
C-type lectin domain family 2, member B
|
|
chr11_+_118443738
|
0.709
|
|
VPS11
|
vacuolar protein sorting 11 homolog (S. cerevisiae)
|
|
chr17_-_54538816
|
0.707
|
|
TRIM37
|
tripartite motif containing 37
|
|
chr9_-_34038869
|
0.705
|
|
UBAP2
|
ubiquitin associated protein 2
|
|
chr10_+_69314714
|
0.705
|
|
SIRT1
|
sirtuin 1
|
|
chr12_-_122415708
|
0.703
|
|
SBNO1
|
strawberry notch homolog 1 (Drosophila)
|
|
chr14_-_19992809
|
0.697
|
|
OSGEP
|
O-sialoglycoprotein endopeptidase
|
|
chr8_-_22145462
|
0.697
|
|
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein
|
|
chr5_+_122138677
|
0.696
|
|
SNX2
|
sorting nexin 2
|
|
chr8_+_17824781
|
0.695
|
|
PCM1
|
pericentriolar material 1
|