|
chr11_-_5110383
|
6.897
|
NM_001005160
|
OR52A5
|
olfactory receptor, family 52, subfamily A, member 5
|
|
chr19_-_12858956
|
6.814
|
NM_006563
|
KLF1
|
Kruppel-like factor 1 (erythroid)
|
|
chr1_-_209818342
|
5.889
|
|
SLC30A1
|
solute carrier family 30 (zinc transporter), member 1
|
|
chr2_+_149119029
|
4.729
|
NM_015630
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila)
|
|
chr1_+_166172420
|
4.545
|
NM_001017977
NM_001198956
NM_001198957
NM_018442
|
DCAF6
|
DDB1 and CUL4 associated factor 6
|
|
chr18_+_9904249
|
4.456
|
|
VAPA
|
VAMP (vesicle-associated membrane protein)-associated protein A, 33kDa
|
|
chr18_+_9904202
|
4.280
|
|
VAPA
|
VAMP (vesicle-associated membrane protein)-associated protein A, 33kDa
|
|
chr17_-_39822366
|
4.170
|
NM_000419
|
ITGA2B
|
integrin, alpha 2b (platelet glycoprotein IIb of IIb/IIIa complex, antigen CD41)
|
|
chr1_-_209818576
|
3.954
|
|
SLC30A1
|
solute carrier family 30 (zinc transporter), member 1
|
|
chr20_+_30160952
|
3.833
|
NM_014742
|
TM9SF4
|
transmembrane 9 superfamily protein member 4
|
|
chr2_+_170074457
|
3.771
|
NM_006063
|
KBTBD10
|
kelch repeat and BTB (POZ) domain containing 10
|
|
chr12_-_69317441
|
3.752
|
NM_001109754
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B
|
|
chr15_+_91244563
|
3.695
|
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr2_+_149118791
|
3.661
|
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila)
|
|
chr10_-_69762654
|
3.649
|
NM_001033083
NM_022129
|
PBLD
|
phenazine biosynthesis-like protein domain containing
|
|
chr11_-_123816190
|
3.600
|
NM_012378
|
OR8B8
|
olfactory receptor, family 8, subfamily B, member 8
|
|
chr7_-_140270749
|
3.535
|
NM_004333
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B1
|
|
chr20_-_33793507
|
3.525
|
|
RBM39
|
RNA binding motif protein 39
|
|
chr17_+_23394062
|
3.509
|
|
NLK
|
nemo-like kinase
|
|
chr1_+_166172545
|
3.494
|
|
DCAF6
|
DDB1 and CUL4 associated factor 6
|
|
chr7_+_104441730
|
3.423
|
NM_018682
NM_182931
|
MLL5
|
myeloid/lymphoid or mixed-lineage leukemia 5 (trithorax homolog, Drosophila)
|
|
chr1_+_22107137
|
3.420
|
|
RPL21
|
ribosomal protein L21
|
|
chr1_+_166172679
|
3.353
|
|
DCAF6
|
DDB1 and CUL4 associated factor 6
|
|
chr6_+_105511615
|
3.329
|
NM_001004317
|
LIN28B
|
lin-28 homolog B (C. elegans)
|
|
chr17_+_72244224
|
3.297
|
|
MFSD11
|
major facilitator superfamily domain containing 11
|
|
chr7_-_150283794
|
3.239
|
NM_172057
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2
|
|
chr16_+_74239135
|
3.220
|
NM_018975
|
TERF2IP
|
telomeric repeat binding factor 2, interacting protein
|
|
chr16_-_30841938
|
3.218
|
|
NCRNA00095
|
non-protein coding RNA 95
|
|
chr11_+_5466490
|
3.072
|
NM_001005163
|
OR52D1
|
olfactory receptor, family 52, subfamily D, member 1
|
|
chr4_-_174774368
|
3.058
|
NM_006792
|
MORF4
|
mortality factor 4
|
|
chr3_-_12680329
|
3.024
|
|
RAF1
|
v-raf-1 murine leukemia viral oncogene homolog 1
|
|
chr10_-_15237351
|
3.023
|
|
LOC100192204
|
peptidylprolyl isomerase A pseudogene
|
|
chrX_-_129347101
|
2.971
|
NM_178471
|
GPR119
|
G protein-coupled receptor 119
|
|
chr1_-_152431175
|
2.919
|
NM_152263
|
TPM3
|
tropomyosin 3
|
|
chr1_+_93070202
|
2.900
|
|
RPL5
|
ribosomal protein L5
|
|
chr5_-_180603335
|
2.873
|
|
GNB2L1
|
guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1
|
|
chr8_+_125556374
|
2.849
|
|
RNF139
|
ring finger protein 139
|
|
chrX_+_48341066
|
2.843
|
|
WDR13
|
WD repeat domain 13
|
|
chr1_-_209818675
|
2.838
|
NM_021194
|
SLC30A1
|
solute carrier family 30 (zinc transporter), member 1
|
|
chr12_+_56135353
|
2.835
|
NM_031479
|
INHBE
|
inhibin, beta E
|
|
chr13_-_107317460
|
2.821
|
NM_001080396
|
FAM155A
|
family with sequence similarity 155, member A
|
|
chr3_-_12680612
|
2.802
|
|
RAF1
|
v-raf-1 murine leukemia viral oncogene homolog 1
|
|
chr14_-_22574193
|
2.799
|
NM_001144932
NM_002797
NM_001130725
|
PSMB5
|
proteasome (prosome, macropain) subunit, beta type, 5
|
|
chr1_+_93070186
|
2.798
|
|
RPL5
|
ribosomal protein L5
|
|
chr7_+_148523486
|
2.780
|
NM_003575
|
ZNF282
|
zinc finger protein 282
|
|
chr15_+_91244422
|
2.772
|
NM_001042572
NM_001271
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr4_+_134293171
|
2.760
|
|
PCDH10
|
protocadherin 10
|
|
chr20_-_33793286
|
2.737
|
|
RBM39
|
RNA binding motif protein 39
|
|
chrX_-_24600661
|
2.732
|
NM_001163264
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr10_+_79463624
|
2.729
|
|
RPS24
|
ribosomal protein S24
|
|
chr3_+_129682434
|
2.712
|
|
|
|
|
chr16_+_3345889
|
2.698
|
NM_012368
|
OR2C1
|
olfactory receptor, family 2, subfamily C, member 1
|
|
chr1_-_243093833
|
2.695
|
|
HNRNPU
|
heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A)
|
|
chr18_+_570356
|
2.691
|
NM_004066
|
CETN1
|
centrin, EF-hand protein, 1
|
|
chr1_+_221966951
|
2.682
|
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr10_+_79463523
|
2.678
|
NM_001026
NM_001142282
NM_001142283
NM_001142284
NM_001142285
NM_033022
|
RPS24
|
ribosomal protein S24
|
|
chrX_-_152938528
|
2.677
|
NM_001025242
NM_001025243
NM_001569
|
IRAK1
|
interleukin-1 receptor-associated kinase 1
|
|
chr1_+_93070120
|
2.625
|
NM_000969
|
RPL5
|
ribosomal protein L5
|
|
chr3_+_142939827
|
2.600
|
|
RNF7
|
ring finger protein 7
|
|
chr2_+_177965734
|
2.599
|
|
AGPS
|
alkylglycerone phosphate synthase
|
|
chr17_-_72244231
|
2.596
|
|
|
|
|
chr1_-_78217360
|
2.587
|
NM_003902
|
FUBP1
|
far upstream element (FUSE) binding protein 1
|
|
chr21_-_33836994
|
2.576
|
NM_001136005
NM_001136006
|
GART
|
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase
|
|
chr7_-_44195405
|
2.565
|
NM_000162
|
GCK
|
glucokinase (hexokinase 4)
|
|
chr9_-_130124820
|
2.563
|
|
TRUB2
|
TruB pseudouridine (psi) synthase homolog 2 (E. coli)
|
|
chr6_+_32003380
|
2.557
|
|
C2
|
complement component 2
|
|
chr2_-_70329067
|
2.553
|
NM_022037
NM_022173
|
TIA1
|
TIA1 cytotoxic granule-associated RNA binding protein
|
|
chr11_-_68505030
|
2.543
|
NM_198923
|
MRGPRD
|
MAS-related GPR, member D
|
|
chr1_+_221966849
|
2.536
|
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr10_+_70745615
|
2.535
|
NM_033496
|
HK1
|
hexokinase 1
|
|
chr10_+_28862068
|
2.518
|
|
|
|
|
chr2_+_218929822
|
2.518
|
NM_198559
|
C2orf62
|
chromosome 2 open reading frame 62
|
|
chr2_+_177965617
|
2.514
|
NM_003659
|
AGPS
|
alkylglycerone phosphate synthase
|
|
chr21_+_33837215
|
2.497
|
NM_032195
NM_138927
|
SON
|
SON DNA binding protein
|
|
chr21_+_33837212
|
2.496
|
|
SON
|
SON DNA binding protein
|
|
chr2_+_114364306
|
2.483
|
|
|
|
|
chr1_-_78217278
|
2.482
|
|
FUBP1
|
far upstream element (FUSE) binding protein 1
|
|
chr1_+_220952518
|
2.480
|
NM_144695
|
BROX
|
BRO1 domain and CAAX motif containing
|
|
chr3_-_12680626
|
2.473
|
NM_002880
|
RAF1
|
v-raf-1 murine leukemia viral oncogene homolog 1
|
|
chr10_+_28862429
|
2.472
|
NM_016628
NM_100486
|
WAC
|
WW domain containing adaptor with coiled-coil
|
|
chr6_+_33047952
|
2.467
|
|
BRD2
|
bromodomain containing 2
|
|
chr17_+_53761293
|
2.453
|
|
LOC100506779
|
hypothetical LOC100506779
|
|
chr17_-_77093468
|
2.446
|
|
ACTG1
|
actin, gamma 1
|
|
chr1_+_221966835
|
2.439
|
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr19_+_6723801
|
2.437
|
|
|
|
|
chr6_-_74284522
|
2.435
|
|
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1
|
|
chr17_+_23393744
|
2.427
|
NM_016231
|
NLK
|
nemo-like kinase
|
|
chr17_-_44037332
|
2.414
|
NM_018952
|
HOXB6
|
homeobox B6
|
|
chr20_-_48980858
|
2.404
|
NM_015339
NM_181442
|
ADNP
|
activity-dependent neuroprotector homeobox
|
|
chr8_+_125556130
|
2.384
|
NM_007218
|
RNF139
|
ring finger protein 139
|
|
chr3_-_187138342
|
2.383
|
|
TRA2B
|
transformer 2 beta homolog (Drosophila)
|
|
chr11_-_64302440
|
2.377
|
|
SF1
|
splicing factor 1
|
|
chr1_-_245942679
|
2.375
|
NM_001005286
|
OR6F1
|
olfactory receptor, family 6, subfamily F, member 1
|
|
chr14_+_28306633
|
2.372
|
|
FOXG1
|
forkhead box G1
|
|
chr21_+_33837233
|
2.364
|
|
SON
|
SON DNA binding protein
|
|
chr8_+_125556294
|
2.362
|
|
RNF139
|
ring finger protein 139
|
|
chr21_-_38792173
|
2.358
|
NM_001136155
NM_182918
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr9_+_130124873
|
2.325
|
|
COQ4
|
coenzyme Q4 homolog (S. cerevisiae)
|
|
chr17_-_36915329
|
2.325
|
NM_002274
NM_153490
|
KRT13
|
keratin 13
|
|
chr5_+_179092405
|
2.322
|
NM_014757
|
MAML1
|
mastermind-like 1 (Drosophila)
|
|
chr21_+_37661728
|
2.318
|
NM_101395
|
DYRK1A
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A
|
|
chr2_-_177965626
|
2.302
|
|
LOC100130691
|
hypothetical LOC100130691
|
|
chr2_+_131203112
|
2.293
|
NM_207364
|
GPR148
|
G protein-coupled receptor 148
|
|
chr17_+_53602013
|
2.289
|
NM_001004707
|
OR4D2
|
olfactory receptor, family 4, subfamily D, member 2
|
|
chr22_+_19601940
|
2.284
|
|
CRKL
|
v-crk sarcoma virus CT10 oncogene homolog (avian)-like
|
|
chr1_-_12830728
|
2.270
|
NM_001146181
|
HNRNPCL1
LOC649330
|
heterogeneous nuclear ribonucleoprotein C-like 1
heterogeneous nuclear ribonucleoprotein C-like
|
|
chr1_-_218168137
|
2.262
|
|
SLC30A10
|
solute carrier family 30, member 10
|
|
chr17_+_55052065
|
2.254
|
|
CLTC
|
clathrin, heavy chain (Hc)
|
|
chr17_-_70690688
|
2.233
|
NM_001005849
NM_006937
|
SUMO2
|
SMT3 suppressor of mif two 3 homolog 2 (S. cerevisiae)
|
|
chr17_+_55052185
|
2.229
|
|
|
|
|
chr9_-_100510657
|
2.228
|
NM_005458
|
GABBR2
|
gamma-aminobutyric acid (GABA) B receptor, 2
|
|
chr5_-_180603476
|
2.220
|
NM_006098
|
GNB2L1
|
guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1
|
|
chr1_-_160648429
|
2.199
|
NM_053282
|
SH2D1B
|
SH2 domain containing 1B
|
|
chr12_-_70343790
|
2.187
|
|
ZFC3H1
|
zinc finger, C3H1-type containing
|
|
chr20_-_33793591
|
2.181
|
|
RBM39
|
RNA binding motif protein 39
|
|
chr9_+_130491644
|
2.174
|
|
SET
|
SET nuclear oncogene
|
|
chr11_+_5558682
|
2.173
|
NM_001005162
|
OR52B6
|
olfactory receptor, family 52, subfamily B, member 6
|
|
chr17_-_70690625
|
2.167
|
|
SUMO2
|
SMT3 suppressor of mif two 3 homolog 2 (S. cerevisiae)
|
|
chr3_+_31549478
|
2.166
|
NM_178862
|
STT3B
|
STT3, subunit of the oligosaccharyltransferase complex, homolog B (S. cerevisiae)
|
|
chr8_+_77928372
|
2.155
|
|
ZFHX4
|
zinc finger homeobox 4
|
|
chr12_-_121577410
|
2.152
|
NM_023012
|
RSRC2
|
arginine/serine-rich coiled-coil 2
|
|
chr22_-_20551953
|
2.143
|
NM_002745
NM_138957
|
MAPK1
|
mitogen-activated protein kinase 1
|
|
chr1_-_146569159
|
2.134
|
NM_001164261
NM_001164262
NM_001144032
|
PPIAL4D
PPIAL4F
PPIAL4E
|
peptidylprolyl isomerase A (cyclophilin A)-like 4D
peptidylprolyl isomerase A (cyclophilin A)-like 4F
peptidylprolyl isomerase A (cyclophilin A)-like 4E
|
|
chr1_+_166516901
|
2.127
|
NM_005149
|
TBX19
|
T-box 19
|
|
chr22_-_20551722
|
2.126
|
|
MAPK1
|
mitogen-activated protein kinase 1
|
|
chr3_+_142939725
|
2.111
|
NM_014245
NM_183237
|
RNF7
|
ring finger protein 7
|
|
chr3_+_142939800
|
2.103
|
|
RNF7
|
ring finger protein 7
|
|
chr17_-_77093603
|
2.101
|
|
ACTG1
|
actin, gamma 1
|
|
chr1_-_220952276
|
2.097
|
NM_022831
|
AIDA
|
axin interactor, dorsalization associated
|
|
chrX_-_152938456
|
2.085
|
|
IRAK1
|
interleukin-1 receptor-associated kinase 1
|
|
chr11_-_123918759
|
2.085
|
NM_001005195
|
OR8B12
|
olfactory receptor, family 8, subfamily B, member 12
|
|
chr3_+_31549435
|
2.070
|
|
STT3B
|
STT3, subunit of the oligosaccharyltransferase complex, homolog B (S. cerevisiae)
|
|
chr17_-_77093709
|
2.063
|
|
ACTG1
|
actin, gamma 1
|
|
chr6_-_2696152
|
2.059
|
NM_001012418
|
MYLK4
|
myosin light chain kinase family, member 4
|
|
chr17_-_70690664
|
2.046
|
|
SUMO2
|
SMT3 suppressor of mif two 3 homolog 2 (S. cerevisiae)
|
|
chr7_-_139123906
|
2.045
|
NM_001113239
NM_022740
|
HIPK2
|
homeodomain interacting protein kinase 2
|
|
chr19_+_10392128
|
2.039
|
NM_001111307
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr17_-_59817722
|
2.022
|
NM_000442
|
PECAM1
|
platelet/endothelial cell adhesion molecule
|
|
chr4_-_83514160
|
2.017
|
NM_001003810
NM_002138
NM_031369
NM_031370
|
HNRNPD
|
heterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37kDa)
|
|
chr10_+_28862514
|
2.013
|
|
WAC
|
WW domain containing adaptor with coiled-coil
|
|
chr18_+_9904032
|
2.010
|
|
VAPA
|
VAMP (vesicle-associated membrane protein)-associated protein A, 33kDa
|
|
chr1_+_221966684
|
2.009
|
NM_001748
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr1_+_35507197
|
2.008
|
|
ZMYM4
|
zinc finger, MYM-type 4
|
|
chr3_+_199161446
|
2.005
|
NM_000996
|
RPL35A
|
ribosomal protein L35a
|
|
chr14_+_19780737
|
1.998
|
NM_001004479
|
OR11H4
|
olfactory receptor, family 11, subfamily H, member 4
|
|
chr17_+_55052046
|
1.997
|
|
CLTC
|
clathrin, heavy chain (Hc)
|
|
chr1_-_42978320
|
1.992
|
NM_001123395
NM_001185117
NM_148960
|
CLDN19
|
claudin 19
|
|
chr2_-_171725150
|
1.992
|
|
TLK1
|
tousled-like kinase 1
|
|
chr1_-_233558071
|
1.990
|
|
ARID4B
|
AT rich interactive domain 4B (RBP1-like)
|
|
chr19_-_52307642
|
1.988
|
|
|
|
|
chr2_-_55698217
|
1.988
|
NM_001122964
NM_020463
|
SMEK2
|
SMEK homolog 2, suppressor of mek1 (Dictyostelium)
|
|
chr1_-_233558114
|
1.985
|
|
ARID4B
|
AT rich interactive domain 4B (RBP1-like)
|
|
chr6_-_88468368
|
1.981
|
|
AKIRIN2
|
akirin 2
|
|
chr1_+_78217510
|
1.979
|
|
DNAJB4
|
DnaJ (Hsp40) homolog, subfamily B, member 4
|
|
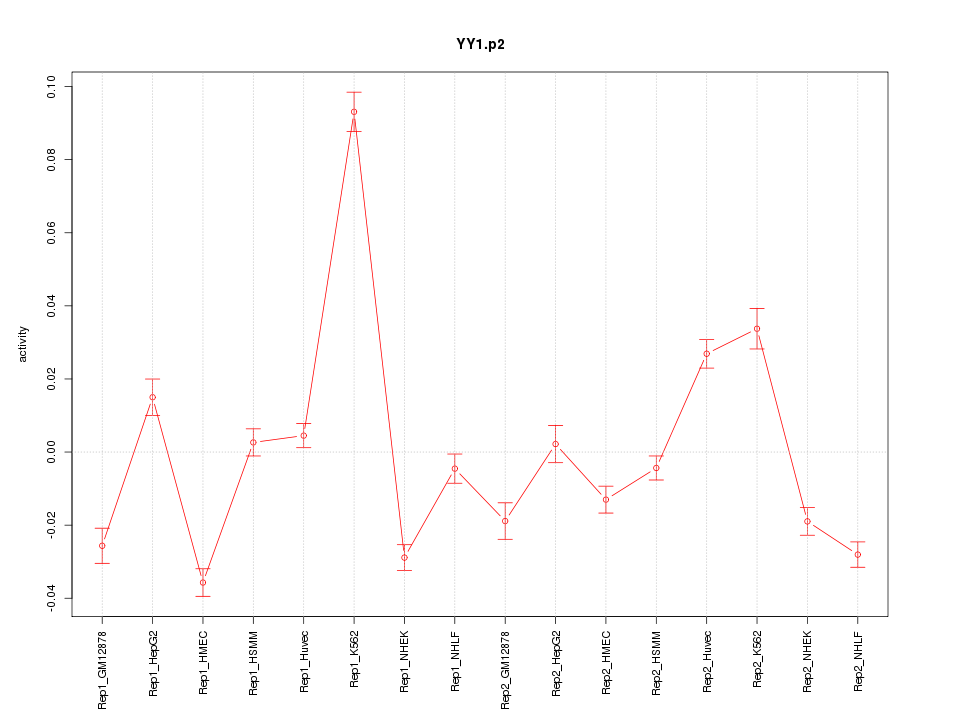
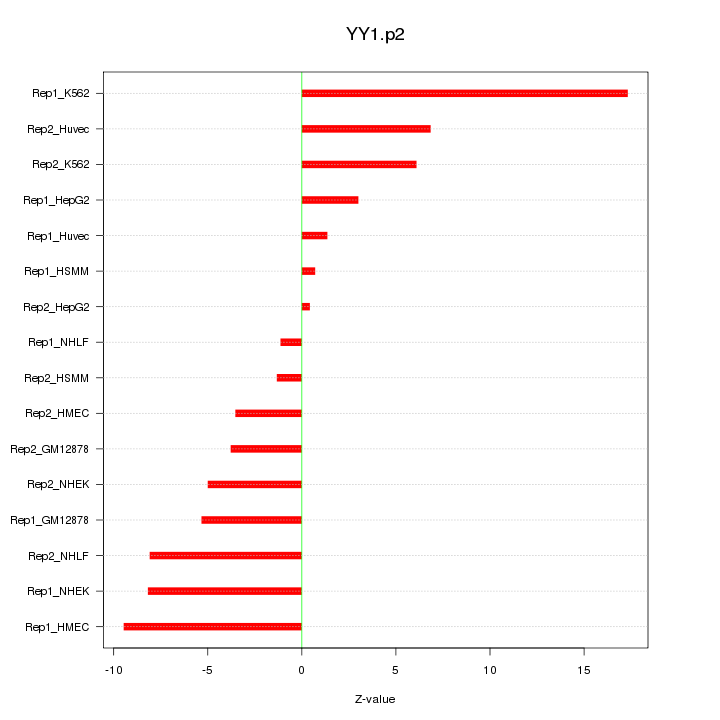
chr14_+_99775185
|
1.976
|
|
YY1
|
YY1 transcription factor
|
|
chr2_+_105224631
|
1.966
|
NM_007227
|
GPR45
|
G protein-coupled receptor 45
|
|
chr2_-_1905508
|
1.962
|
|
MYT1L
|
myelin transcription factor 1-like
|
|
chr2_+_236067687
|
1.957
|
|
AGAP1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1
|
|
chr13_+_49487390
|
1.956
|
NM_173605
NM_199464
|
TRIM13
KCNRG
|
tripartite motif containing 13
potassium channel regulator
|
|
chr20_+_30161173
|
1.950
|
|
TM9SF4
|
transmembrane 9 superfamily protein member 4
|
|
chr5_+_180726874
|
1.949
|
NM_001005224
NM_001005221
NM_001005277
|
OR4F3
OR4F29
OR4F16
|
olfactory receptor, family 4, subfamily F, member 3
olfactory receptor, family 4, subfamily F, member 29
olfactory receptor, family 4, subfamily F, member 16
|
|
chr12_+_54672766
|
1.949
|
|
RAB5B
|
RAB5B, member RAS oncogene family
|
|
chr16_+_69115227
|
1.945
|
|
SF3B3
|
splicing factor 3b, subunit 3, 130kDa
|
|
chr1_-_27113018
|
1.945
|
NM_021969
|
NR0B2
|
nuclear receptor subfamily 0, group B, member 2
|
|
chr16_-_86460529
|
1.941
|
NM_003486
|
SLC7A5
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 5
|
|
chr2_+_182464910
|
1.941
|
|
SSFA2
|
sperm specific antigen 2
|
|
chr11_-_64302509
|
1.940
|
NM_001178030
|
SF1
|
splicing factor 1
|
|
chr2_+_3600984
|
1.934
|
|
RPS7
|
ribosomal protein S7
|
|
chr15_-_100280784
|
1.933
|
NM_001004195
|
OR4F4
OR4F17
|
olfactory receptor, family 4, subfamily F, member 4
olfactory receptor, family 4, subfamily F, member 17
|
|
chr1_+_220952681
|
1.932
|
|
BROX
|
BRO1 domain and CAAX motif containing
|
|
chr8_-_107023
|
1.925
|
NM_001005504
|
OR4F21
|
olfactory receptor, family 4, subfamily F, member 21
|
|
chrX_-_152938441
|
1.922
|
|
IRAK1
|
interleukin-1 receptor-associated kinase 1
|
|
chr15_+_57286313
|
1.913
|
NM_033195
|
LDHAL6B
|
lactate dehydrogenase A-like 6B
|
|
chr10_-_61819493
|
1.910
|
NM_020987
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr17_-_53764867
|
1.906
|
|
|
|
|
chr1_-_46541408
|
1.905
|
|
LRRC41
|
leucine rich repeat containing 41
|
|
chr8_+_10567556
|
1.902
|
NM_001040032
|
C8orf74
|
chromosome 8 open reading frame 74
|
|
chr19_+_13736345
|
1.900
|
|
MRI1
|
methylthioribose-1-phosphate isomerase homolog (S. cerevisiae)
|
|
chr1_-_46541439
|
1.900
|
|
LRRC41
|
leucine rich repeat containing 41
|
|
chr1_+_51208229
|
1.898
|
NM_078626
|
CDKN2C
|
cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4)
|
|
chr13_-_36577800
|
1.898
|
NM_145203
|
CSNK1A1L
|
casein kinase 1, alpha 1-like
|
|
chr8_+_145692916
|
1.895
|
NM_032902
|
PPP1R16A
|
protein phosphatase 1, regulatory (inhibitor) subunit 16A
|
|
chr1_+_51208499
|
1.892
|
|
CDKN2C
|
cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4)
|
|
chr1_-_233558117
|
1.891
|
NM_016374
NM_031371
|
ARID4B
|
AT rich interactive domain 4B (RBP1-like)
|
|
chr17_+_55052060
|
1.891
|
|
CLTC
|
clathrin, heavy chain (Hc)
|
|
chr2_+_104838177
|
1.880
|
NM_006236
|
POU3F3
|
POU class 3 homeobox 3
|
|
chr22_+_43451600
|
1.876
|
|
PRR5
|
proline rich 5 (renal)
|
|
chr14_-_20340377
|
1.872
|
NM_198232
|
RNASE1
|
ribonuclease, RNase A family, 1 (pancreatic)
|
|
chr15_+_32425357
|
1.869
|
NM_175741
|
C15orf55
|
chromosome 15 open reading frame 55
|
|
chr1_-_218168404
|
1.866
|
|
|
|
|
chr9_-_124370576
|
1.865
|
NM_001004454
|
OR1L8
|
olfactory receptor, family 1, subfamily L, member 8
|
|
chr19_+_5574119
|
1.863
|
|
SAFB
|
scaffold attachment factor B
|
|
chrX_+_48340859
|
1.863
|
|
WDR13
|
WD repeat domain 13
|
|
chr18_+_9904060
|
1.859
|
|
VAPA
|
VAMP (vesicle-associated membrane protein)-associated protein A, 33kDa
|
|
chr3_+_180805266
|
1.847
|
NM_002492
|
NDUFB5
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16kDa
|
|
chr7_-_26206845
|
1.838
|
|
HNRNPA2B1
|
heterogeneous nuclear ribonucleoprotein A2/B1
|
|
chr1_+_229181417
|
1.829
|
NM_022786
|
ARV1
|
ARV1 homolog (S. cerevisiae)
|
|
chr5_+_10303377
|
1.827
|
|
CCT5
|
chaperonin containing TCP1, subunit 5 (epsilon)
|
|
chr3_-_115825742
|
1.826
|
NM_001164347
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr3_+_44992733
|
1.813
|
NM_015004
|
EXOSC7
|
exosome component 7
|