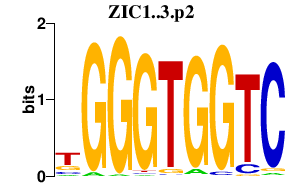
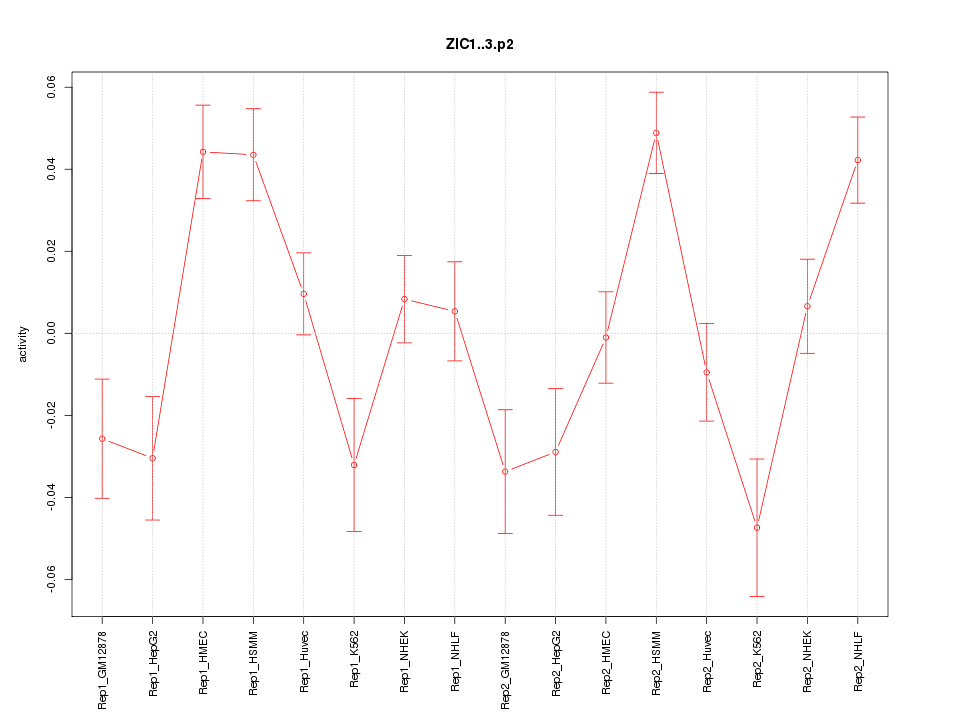
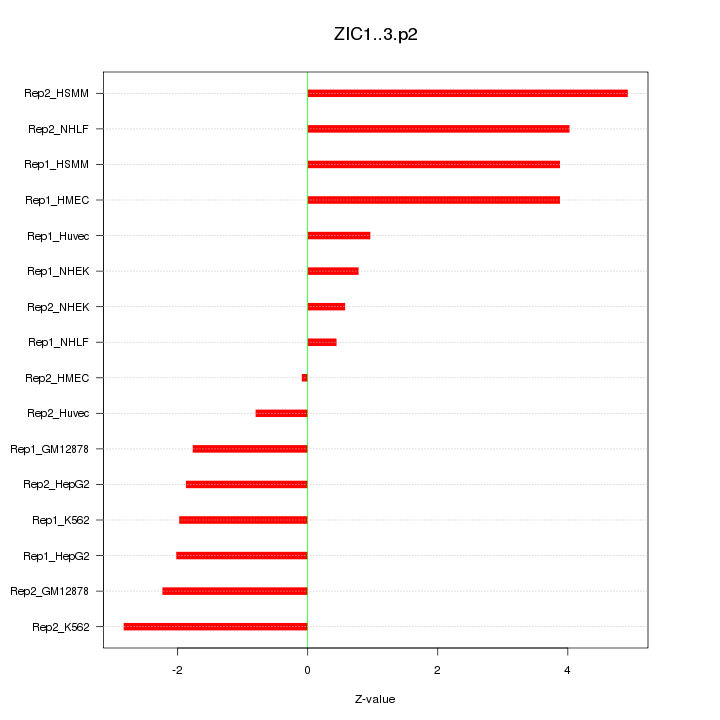
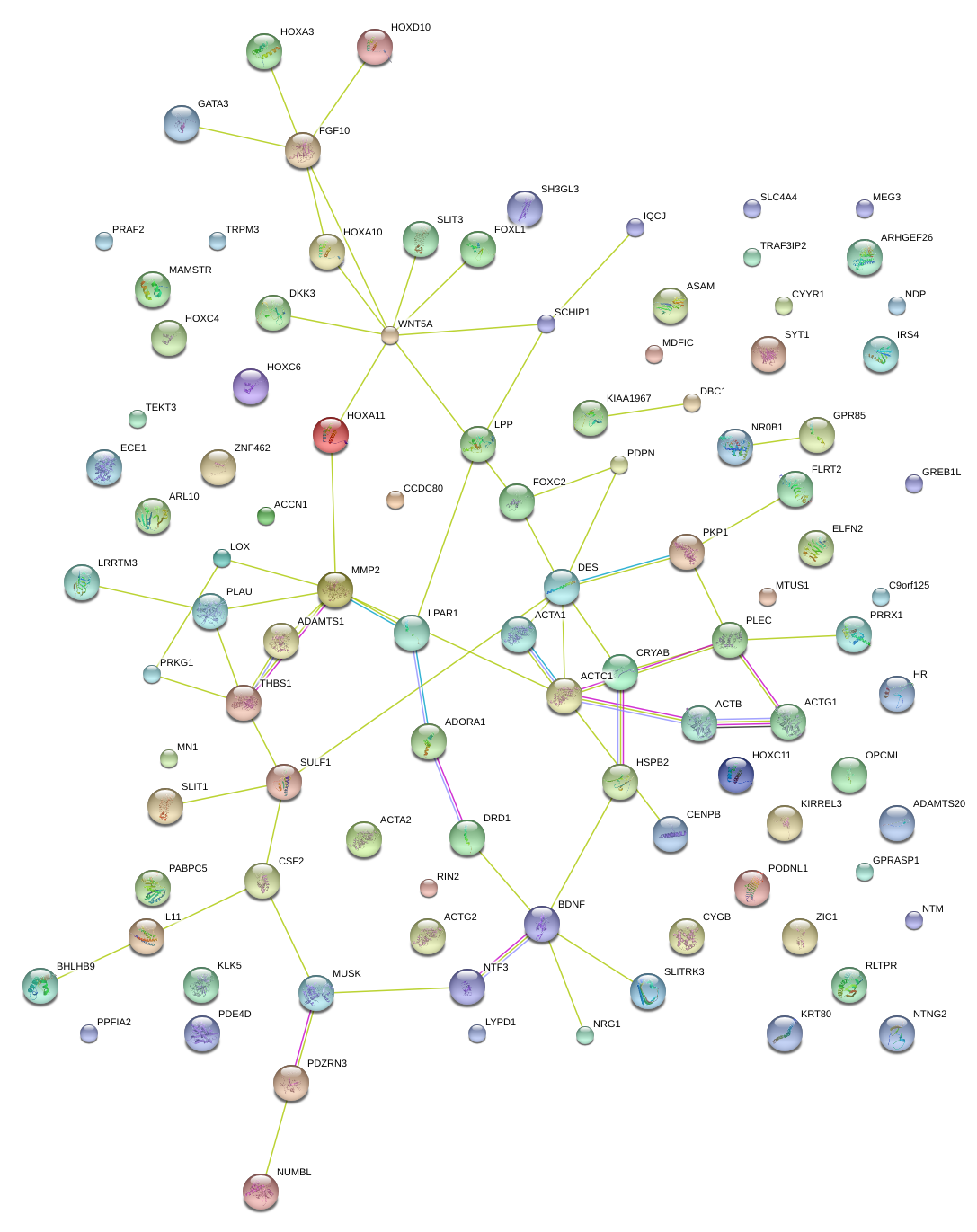
|
chr3_-_73756668
|
8.134
|
NM_015009
|
PDZRN3
|
PDZ domain containing ring finger 3
|
|
chr16_+_85169615
|
5.428
|
NM_005250
|
FOXL1
|
forkhead box L1
|
|
chr3_+_64645819
|
5.260
|
|
LOC100507098
|
hypothetical LOC100507098
|
|
chr21_-_26867138
|
4.886
|
NM_052954
|
CYYR1
|
cysteine/tyrosine-rich 1
|
|
chr10_-_90702490
|
4.536
|
NM_001613
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta
|
|
chr7_-_27191287
|
3.656
|
NM_005523
|
HOXA11
|
homeobox A11
|
|
chr12_+_52653176
|
3.528
|
NM_014212
|
HOXC11
|
homeobox C11
|
|
chr5_-_168660293
|
3.381
|
NM_003062
|
SLIT3
|
slit homolog 3 (Drosophila)
|
|
chr10_+_75343352
|
3.291
|
|
PLAU
|
plasminogen activator, urokinase
|
|
chr22_-_36153449
|
3.037
|
NM_052906
|
ELFN2
|
extracellular leucine-rich repeat and fibronectin type III domain containing 2
|
|
chr20_+_19903778
|
2.924
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chr11_-_126375864
|
2.922
|
|
KIRREL3
|
kin of IRRE like 3 (Drosophila)
|
|
chr4_+_72271218
|
2.902
|
NM_001098484
NM_001134742
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr14_+_85066265
|
2.869
|
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr19_-_13910167
|
2.780
|
NM_024825
|
PODNL1
|
podocan-like 1
|
|
chr1_+_199519202
|
2.755
|
NM_000299
NM_001005337
|
PKP1
|
plakophilin 1 (ectodermal dysplasia/skin fragility syndrome)
|
|
chr21_-_27139143
|
2.669
|
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1
|
|
chr21_-_27139563
|
2.624
|
NM_006988
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1
|
|
chr9_+_108665168
|
2.623
|
NM_021224
|
ZNF462
|
zinc finger protein 462
|
|
chr22_-_26527469
|
2.585
|
NM_002430
|
MN1
|
meningioma (disrupted in balanced translocation) 1
|
|
chr17_-_15185623
|
2.568
|
NM_031898
|
TEKT3
|
tektin 3
|
|
chr11_-_126375895
|
2.547
|
NM_001161707
NM_032531
|
KIRREL3
|
kin of IRRE like 3 (Drosophila)
|
|
chr5_+_131437383
|
2.535
|
NM_000758
|
CSF2
|
colony stimulating factor 2 (granulocyte-macrophage)
|
|
chr9_-_72673753
|
2.513
|
NM_001007470
NM_020952
NM_024971
NM_206944
NM_206945
NM_206946
NM_206947
NM_206948
|
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3
|
|
chr11_+_131285747
|
2.470
|
NM_001144058
NM_001144059
NM_016522
|
NTM
|
neurotrimin
|
|
chr12_-_50871953
|
2.418
|
NM_001081492
NM_182507
|
KRT80
|
keratin 80
|
|
chrX_+_90576252
|
2.395
|
NM_080832
|
PABPC5
|
poly(A) binding protein, cytoplasmic 5
|
|
chr10_+_52421120
|
2.377
|
|
PRKG1
|
protein kinase, cGMP-dependent, type I
|
|
chr16_+_85158357
|
2.364
|
NM_005251
|
FOXC2
|
forkhead box C2 (MFH-1, mesenchyme forkhead 1)
|
|
chr5_+_175725050
|
2.339
|
NM_173664
|
ARL10
|
ADP-ribosylation factor-like 10
|
|
chr11_-_11986704
|
2.305
|
NM_001018057
|
DKK3
|
dickkopf homolog 3 (Xenopus laevis)
|
|
chr7_-_27180398
|
2.287
|
NM_018951
|
HOXA10
|
homeobox A10
|
|
chr12_+_5411534
|
2.277
|
NM_001102654
|
NTF3
|
neurotrophin 3
|
|
chr7_+_27191534
|
2.216
|
|
HOXA11-AS1
|
HOXA11 antisense RNA 1 (non-protein coding)
|
|
chr17_-_72045218
|
2.207
|
NM_134268
|
CYGB
|
cytoglobin
|
|
chr2_-_133144094
|
2.201
|
|
LYPD1
|
LY6/PLAUR domain containing 1
|
|
chrX_-_107866262
|
2.198
|
NM_003604
|
IRS4
|
insulin receptor substrate 4
|
|
chr17_-_28644118
|
2.136
|
NM_183377
|
ACCN1
|
amiloride-sensitive cation channel 1, neuronal
|
|
chr19_-_56147971
|
2.120
|
NM_001077491
NM_001077492
NM_012427
|
KLK5
|
kallikrein-related peptidase 5
|
|
chr6_+_101948102
|
2.092
|
|
|
|
|
chr3_+_155321826
|
2.085
|
NM_015595
|
ARHGEF26
|
Rho guanine nucleotide exchange factor (GEF) 26
|
|
chr11_-_111287657
|
2.076
|
NM_001885
|
CRYAB
|
crystallin, alpha B
|
|
chr3_+_189353502
|
2.049
|
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chrX_-_43717864
|
2.044
|
NM_000266
|
NDP
|
Norrie disease (pseudoglioma)
|
|
chr11_+_111288669
|
2.037
|
NM_001541
|
HSPB2-C11ORF52
HSPB2
|
HSPB2-C11orf52 read-through transcript
heat shock 27kDa protein 2
|
|
chr9_+_112470871
|
2.032
|
NM_001166280
NM_001166281
NM_005592
|
MUSK
|
muscle, skeletal, receptor tyrosine kinase
|
|
chr11_+_111288686
|
1.999
|
|
HSPB2-C11ORF52
HSPB2
|
HSPB2-C11orf52 read-through transcript
heat shock 27kDa protein 2
|
|
chr8_+_31616809
|
1.991
|
NM_013962
|
NRG1
|
neuregulin 1
|
|
chr9_+_134027154
|
1.958
|
NM_032536
|
NTNG2
|
netrin G2
|
|
chr5_-_58607673
|
1.957
|
NM_001197220
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chrX_+_101887562
|
1.954
|
NM_001142529
NM_001142530
|
BHLHB9
|
basic helix-loop-helix domain containing, class B, 9
|
|
chr5_-_44424423
|
1.950
|
NM_004465
|
FGF10
|
fibroblast growth factor 10
|
|
chr7_-_112513379
|
1.939
|
NM_001146266
NM_001146267
|
GPR85
|
G protein-coupled receptor 85
|
|
chr16_+_66244314
|
1.919
|
|
RLTPR
|
RGD motif, leucine rich repeats, tropomodulin domain and proline-rich containing
|
|
chr9_-_112840064
|
1.915
|
NM_001401
NM_057159
|
LPAR1
|
lysophosphatidic acid receptor 1
|
|
chr19_-_60573552
|
1.902
|
NM_000641
|
IL11
|
interleukin 11
|
|
chr11_-_27697768
|
1.873
|
NM_001143807
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr1_+_168899936
|
1.853
|
NM_006902
NM_022716
|
PRRX1
|
paired related homeobox 1
|
|
chr7_+_114350144
|
1.835
|
|
MDFIC
|
MyoD family inhibitor domain containing
|
|
chr9_-_121171400
|
1.833
|
NM_014618
|
DBC1
|
deleted in bladder cancer 1
|
|
chr12_-_42231990
|
1.822
|
NM_025003
|
ADAMTS20
|
ADAM metallopeptidase with thrombospondin type 1 motif, 20
|
|
chr1_+_201363458
|
1.817
|
NM_000674
NM_001048230
|
ADORA1
|
adenosine A1 receptor
|
|
chr5_-_174803768
|
1.805
|
NM_000794
|
DRD1
|
dopamine receptor D1
|
|
chr7_+_27190688
|
1.771
|
|
|
|
|
chr8_-_17599438
|
1.750
|
NM_020749
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chrX_+_101792949
|
1.742
|
NM_001099410
NM_001099411
NM_014710
NM_001184727
|
GPRASP1
|
G protein-coupled receptor associated sorting protein 1
|
|
chr8_+_70541631
|
1.741
|
|
SULF1
|
sulfatase 1
|
|
chr15_+_37660568
|
1.741
|
NM_003246
|
THBS1
|
thrombospondin 1
|
|
chr3_-_166397142
|
1.719
|
NM_014926
|
SLITRK3
|
SLIT and NTRK-like family, member 3
|
|
chrX_-_30237403
|
1.717
|
NM_000475
|
NR0B1
|
nuclear receptor subfamily 0, group B, member 1
|
|
chr3_-_55490401
|
1.696
|
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr6_-_112001590
|
1.693
|
NM_001164282
|
TRAF3IP2
|
TRAF3 interacting protein 2
|
|
chr5_-_121440704
|
1.673
|
NM_001178102
|
LOX
|
lysyl oxidase
|
|
chr12_-_80676562
|
1.660
|
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2
|
|
chr6_-_169392771
|
1.654
|
|
|
|
|
chrX_-_48818585
|
1.643
|
NM_007213
|
PRAF2
|
PRA1 domain family, member 2
|
|
chr19_-_45888259
|
1.640
|
NM_004756
|
NUMBL
|
numb homolog (Drosophila)-like
|
|
chr11_-_122571046
|
1.614
|
|
CLMP
|
CXADR-like membrane protein
|
|
chr2_+_219991342
|
1.601
|
NM_001927
|
DES
|
desmin
|
|
chr7_-_27120015
|
1.598
|
|
HOXA3
|
homeobox A3
|
|
chr9_+_134026836
|
1.588
|
|
NTNG2
|
netrin G2
|
|
chr3_-_113841200
|
1.584
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr8_-_22044463
|
1.580
|
NM_005144
NM_018411
|
HR
|
hairless homolog (mouse)
|
|
chr11_-_132318741
|
1.578
|
|
OPCML
|
opioid binding protein/cell adhesion molecule-like
|
|
chr11_-_110917632
|
1.559
|
|
|
|
|
chr9_-_103289154
|
1.550
|
NM_032342
|
C9orf125
|
chromosome 9 open reading frame 125
|
|
chr20_-_3713509
|
1.544
|
|
CENPB
|
centromere protein B, 80kDa
|
|
chr14_+_100362236
|
1.541
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr12_+_52733927
|
1.539
|
NM_153633
|
HOXC4
HOXC6
|
homeobox C4
homeobox C6
|
|
chr11_-_11987115
|
1.539
|
|
DKK3
|
dickkopf homolog 3 (Xenopus laevis)
|
|
chr1_-_21478571
|
1.534
|
NM_001113347
|
ECE1
|
endothelin converting enzyme 1
|
|
chr4_+_159311353
|
1.530
|
|
LOC285505
|
hypothetical protein LOC285505
|
|
chr18_+_17076166
|
1.526
|
NM_001142966
|
GREB1L
|
growth regulation by estrogen in breast cancer-like
|
|
chr8_-_145097031
|
1.511
|
NM_201380
|
PLEC
|
plectin
|
|
chr10_+_8137356
|
1.508
|
|
GATA3
|
GATA binding protein 3
|
|
chr16_+_54070302
|
1.495
|
NM_004530
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase)
|
|
chr15_+_81906983
|
1.487
|
NM_003027
|
SH3GL3
|
SH3-domain GRB2-like 3
|
|
chr19_-_53912025
|
1.476
|
NM_182574
|
MAMSTR
|
MEF2 activating motif and SAP domain containing transcriptional regulator
|
|
chr14_+_100362204
|
1.467
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr3_+_160964574
|
1.459
|
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr12_+_77782925
|
1.457
|
|
SYT1
|
synaptotagmin I
|
|
chr1_+_13782838
|
1.456
|
NM_006474
NM_198389
|
PDPN
|
podoplanin
|
|
chr10_+_68355797
|
1.443
|
NM_178011
|
LRRTM3
|
leucine rich repeat transmembrane neuronal 3
|
|
chr3_+_148610516
|
1.437
|
|
ZIC1
|
Zic family member 1 (odd-paired homolog, Drosophila)
|
|
chr5_-_83715947
|
1.433
|
|
EDIL3
|
EGF-like repeats and discoidin I-like domains 3
|
|
chr17_+_7549138
|
1.430
|
NM_001406
|
EFNB3
|
ephrin-B3
|
|
chr10_+_106018915
|
1.424
|
|
GSTO2
|
glutathione S-transferase omega 2
|
|
chr20_-_22978141
|
1.417
|
NM_000361
|
THBD
|
thrombomodulin
|
|
chr9_+_111850698
|
1.405
|
NM_001004065
NM_001198656
|
AKAP2
|
A kinase (PRKA) anchor protein 2
|
|
chr16_-_31390926
|
1.395
|
|
|
|
|
chr4_+_166519392
|
1.387
|
NM_001873
|
CPE
|
carboxypeptidase E
|
|
chrX_-_114374860
|
1.381
|
NM_020871
|
LRCH2
|
leucine-rich repeats and calponin homology (CH) domain containing 2
|
|
chr9_+_4480193
|
1.380
|
NM_004170
|
SLC1A1
|
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1
|
|
chr2_+_217206450
|
1.371
|
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa
|
|
chr18_+_54013575
|
1.362
|
NM_001144970
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr3_-_21767819
|
1.361
|
NM_024697
|
ZNF385D
|
zinc finger protein 385D
|
|
chr7_-_27190882
|
1.341
|
|
HOXA11
|
homeobox A11
|
|
chr15_-_98959898
|
1.337
|
NM_001040616
|
LINS
|
lines homolog (Drosophila)
|
|
chr4_+_156808261
|
1.325
|
NM_001130684
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3
|
|
chr20_+_61798512
|
1.315
|
|
TNFRSF6B
|
tumor necrosis factor receptor superfamily, member 6b, decoy
|
|
chr20_+_61798446
|
1.310
|
NM_003823
|
TNFRSF6B
|
tumor necrosis factor receptor superfamily, member 6b, decoy
|
|
chr8_-_93144366
|
1.306
|
NM_004349
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr1_+_234372454
|
1.303
|
NM_003272
|
GPR137B
|
G protein-coupled receptor 137B
|
|
chr12_+_77782719
|
1.300
|
|
SYT1
|
synaptotagmin I
|
|
chr1_+_201364050
|
1.283
|
|
ADORA1
|
adenosine A1 receptor
|
|
chr2_+_217206327
|
1.275
|
NM_000597
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa
|
|
chr9_+_78824390
|
1.264
|
NM_001013735
|
FOXB2
|
forkhead box B2
|
|
chr1_+_116181927
|
1.264
|
|
NHLH2
|
nescient helix loop helix 2
|
|
chr6_+_133604181
|
1.261
|
NM_004100
NM_172103
NM_172105
|
EYA4
|
eyes absent homolog 4 (Drosophila)
|
|
chr12_-_21985433
|
1.259
|
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9
|
|
chr6_+_41714171
|
1.252
|
NM_005586
|
MDFI
|
MyoD family inhibitor
|
|
chr8_+_1909437
|
1.249
|
NM_014867
|
KBTBD11
|
kelch repeat and BTB (POZ) domain containing 11
|
|
chr12_+_54400417
|
1.244
|
NM_002905
|
RDH5
|
retinol dehydrogenase 5 (11-cis/9-cis)
|
|
chr5_+_140835788
|
1.238
|
|
PCDHGC3
|
protocadherin gamma subfamily C, 3
|
|
chr16_+_31390567
|
1.237
|
NM_001042454
NM_001164719
|
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1
|
|
chr13_+_30378311
|
1.235
|
NM_032849
|
C13orf33
|
chromosome 13 open reading frame 33
|
|
chr6_+_26348539
|
1.233
|
NM_003540
|
HIST1H4F
|
histone cluster 1, H4f
|
|
chr1_-_212791188
|
1.227
|
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14
|
|
chr10_+_119292699
|
1.217
|
|
|
|
|
chr15_-_53996597
|
1.216
|
NM_198400
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr22_-_49063088
|
1.214
|
|
PLXNB2
|
plexin B2
|
|
chr10_-_71575953
|
1.202
|
|
TYSND1
|
trypsin domain containing 1
|
|
chr5_+_140326473
|
1.201
|
NM_031883
|
PCDHAC2
|
protocadherin alpha subfamily C, 2
|
|
chr5_-_92982927
|
1.198
|
|
|
|
|
chr12_-_80677223
|
1.190
|
NM_003625
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2
|
|
chr20_+_35583020
|
1.187
|
NM_005386
NM_181689
|
NNAT
|
neuronatin
|
|
chr3_+_3816079
|
1.187
|
NM_020873
|
LRRN1
|
leucine rich repeat neuronal 1
|
|
chr12_+_6179736
|
1.187
|
NM_001769
|
CD9
|
CD9 molecule
|
|
chr12_+_77782648
|
1.187
|
|
SYT1
|
synaptotagmin I
|
|
chr11_+_19691397
|
1.184
|
NM_145117
NM_182964
|
NAV2
|
neuron navigator 2
|
|
chr22_-_42589543
|
1.181
|
NM_014351
|
SULT4A1
|
sulfotransferase family 4A, member 1
|
|
chr8_-_93144039
|
1.179
|
|
|
|
|
chr2_+_220033777
|
1.175
|
NM_001173476
|
SPEG
|
SPEG complex locus
|
|
chr14_+_64077171
|
1.162
|
|
HSPA2
|
heat shock 70kDa protein 2
|
|
chr4_-_90977149
|
1.162
|
NM_001146054
NM_007308
NM_000345
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor)
|
|
chr8_+_75899326
|
1.156
|
NM_015886
|
PI15
|
peptidase inhibitor 15
|
|
chr7_-_150576634
|
1.147
|
|
SMARCD3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3
|
|
chr5_+_140326237
|
1.137
|
|
PCDHAC2
|
protocadherin alpha subfamily C, 2
|
|
chr11_-_35503720
|
1.137
|
NM_001001991
NM_015430
|
PAMR1
|
peptidase domain containing associated with muscle regeneration 1
|
|
chr4_+_15089931
|
1.135
|
|
CC2D2A
|
coiled-coil and C2 domain containing 2A
|
|
chrX_+_51653348
|
1.132
|
NM_001005333
NM_006986
|
MAGED1
|
melanoma antigen family D, 1
|
|
chr17_-_30838870
|
1.125
|
NM_001195790
|
SLFN12L
|
schlafen family member 12-like
|
|
chr11_-_11987198
|
1.121
|
NM_015881
|
DKK3
|
dickkopf homolog 3 (Xenopus laevis)
|
|
chr6_+_142664490
|
1.120
|
NM_001032394
NM_001032395
NM_020455
NM_198569
|
GPR126
|
G protein-coupled receptor 126
|
|
chr11_+_122806196
|
1.103
|
|
LOC100128242
|
hypothetical LOC100128242
|
|
chr19_-_13478037
|
1.103
|
NM_000068
NM_001127221
NM_001127222
NM_001174080
NM_023035
|
CACNA1A
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit
|
|
chr14_+_104218487
|
1.099
|
|
|
|
|
chr6_+_148705817
|
1.094
|
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr7_+_44110527
|
1.091
|
|
AEBP1
|
AE binding protein 1
|
|
chr12_-_131697109
|
1.089
|
NM_001195520
|
LOC645277
|
hypothetical LOC645277
|
|
chr5_+_140835727
|
1.087
|
NM_002588
NM_032402
NM_032403
|
PCDHGC3
|
protocadherin gamma subfamily C, 3
|
|
chr6_-_166321516
|
1.081
|
|
C6orf176
|
chromosome 6 open reading frame 176
|
|
chr19_-_13808101
|
1.078
|
|
LOC284454
|
hypothetical LOC284454
|
|
chr6_+_142665057
|
1.072
|
|
GPR126
|
G protein-coupled receptor 126
|
|
chr7_-_107883929
|
1.063
|
NM_001193582
NM_001193583
NM_001193584
NM_005010
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr4_+_156807311
|
1.052
|
NM_000856
NM_001130682
NM_001130686
NM_001130687
NM_001130683
NM_001130685
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3
|
|
chr11_+_19328846
|
1.052
|
NM_001111018
|
NAV2
|
neuron navigator 2
|
|
chr1_+_232107170
|
1.049
|
NM_173508
|
SLC35F3
|
solute carrier family 35, member F3
|
|
chr15_-_27901828
|
1.046
|
NM_003257
NM_175610
|
TJP1
|
tight junction protein 1 (zona occludens 1)
|
|
chr8_+_76059262
|
1.043
|
NM_031461
|
CRISPLD1
|
cysteine-rich secretory protein LCCL domain containing 1
|
|
chr1_+_17817397
|
1.041
|
|
ARHGEF10L
|
Rho guanine nucleotide exchange factor (GEF) 10-like
|
|
chr1_+_200124412
|
1.038
|
NM_198149
|
SHISA4
|
shisa homolog 4 (Xenopus laevis)
|
|
chr8_-_132122016
|
1.033
|
NM_001115
|
ADCY8
|
adenylate cyclase 8 (brain)
|
|
chr14_+_64076916
|
1.031
|
NM_021979
|
HSPA2
|
heat shock 70kDa protein 2
|
|
chr5_-_134942346
|
1.030
|
|
CXCL14
|
chemokine (C-X-C motif) ligand 14
|
|
chr10_-_105202012
|
1.018
|
|
CALHM2
|
calcium homeostasis modulator 2
|
|
chr7_-_27102049
|
1.016
|
NM_005522
NM_153620
|
HOXA1
|
homeobox A1
|
|
chr5_+_167889123
|
1.016
|
|
|
|
|
chr6_+_17389752
|
1.010
|
NM_001143942
|
RBM24
|
RNA binding motif protein 24
|
|
chr3_+_115099007
|
1.009
|
NM_001172105
|
GRAMD1C
|
GRAM domain containing 1C
|
|
chr1_+_110494654
|
1.008
|
NM_001010898
|
SLC6A17
|
solute carrier family 6, member 17
|
|
chr19_+_10597167
|
1.007
|
NM_020428
|
SLC44A2
|
solute carrier family 44, member 2
|
|
chr8_+_39084206
|
1.006
|
NM_145004
|
ADAM32
|
ADAM metallopeptidase domain 32
|
|
chr8_-_11096257
|
1.005
|
NM_173683
|
XKR6
|
XK, Kell blood group complex subunit-related family, member 6
|
|
chr3_-_113841448
|
1.004
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr3_+_50281582
|
1.003
|
|
SEMA3B
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B
|
|
chr6_+_19946591
|
0.999
|
|
|
|
|
chr4_-_159311959
|
0.992
|
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr2_-_192767844
|
0.991
|
NM_016192
|
TMEFF2
|
transmembrane protein with EGF-like and two follistatin-like domains 2
|
|
chr15_+_98960277
|
0.985
|
NM_024708
NM_198243
|
ASB7
|
ankyrin repeat and SOCS box containing 7
|