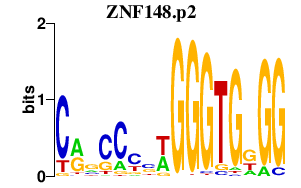
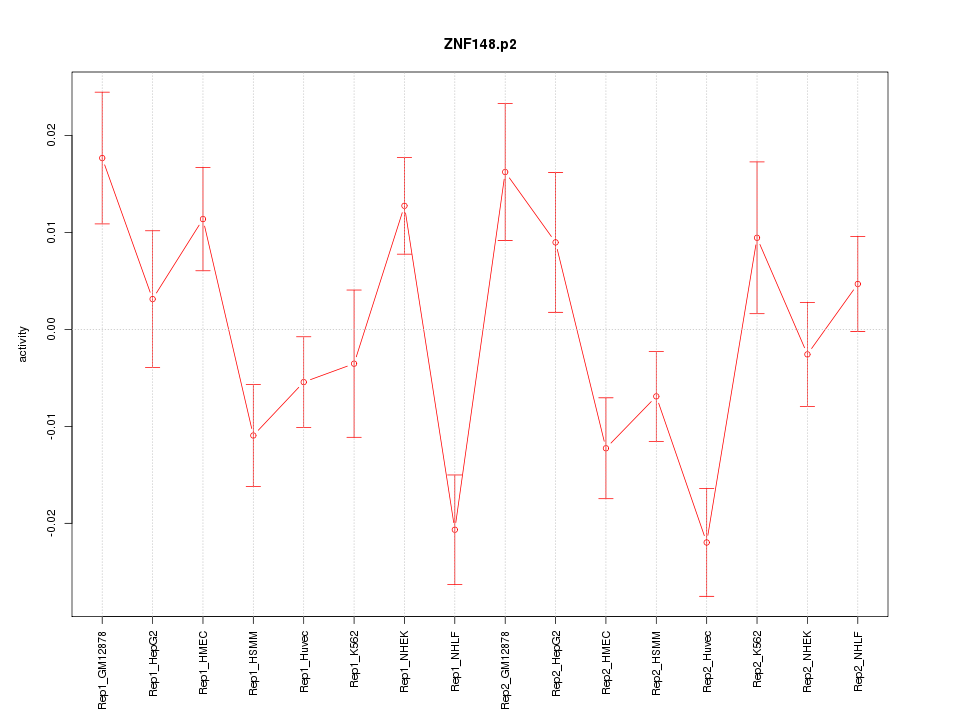
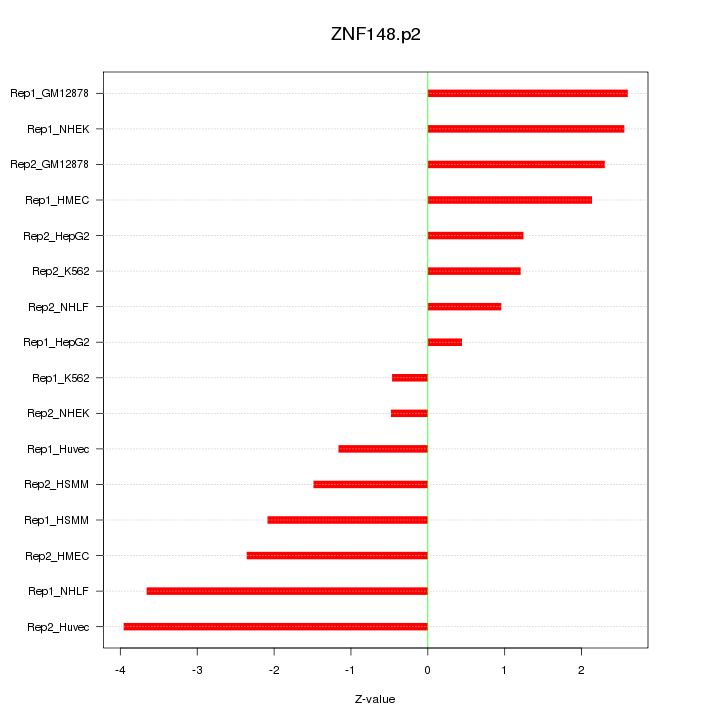
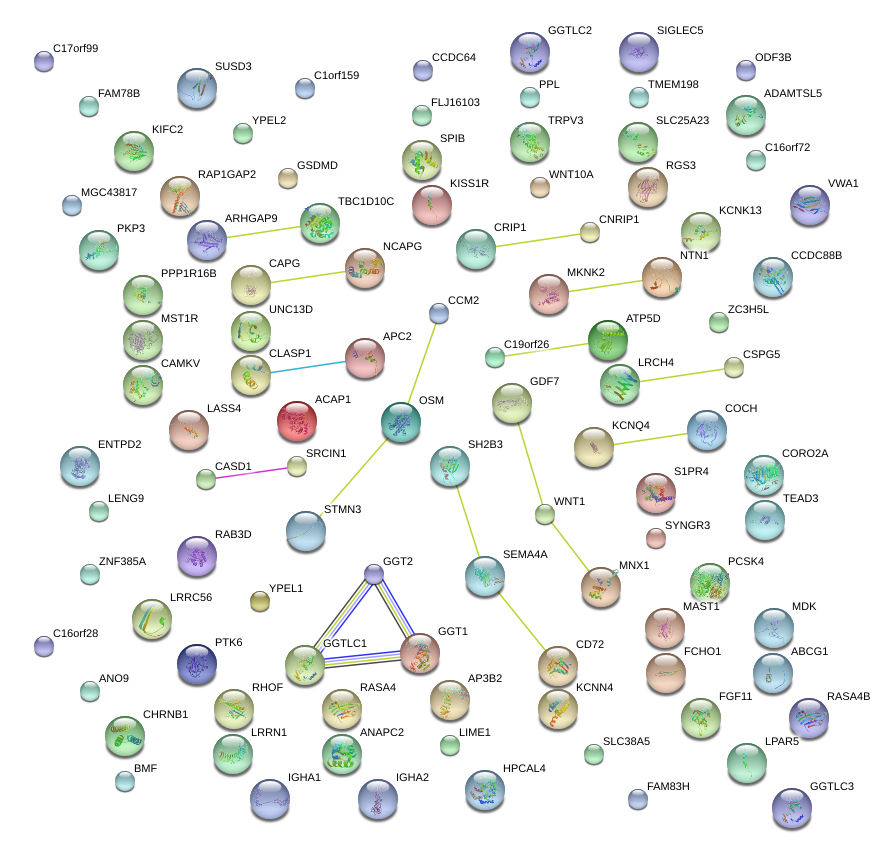
|
chr11_-_431995
|
2.881
|
NM_001012302
|
ANO9
|
anoctamin 9
|
|
chr11_-_431956
|
2.806
|
|
ANO9
|
anoctamin 9
|
|
chr16_+_1979959
|
2.169
|
NM_004209
|
SYNGR3
|
synaptogyrin 3
|
|
chr3_-_49916304
|
2.023
|
NM_002447
|
MST1R
|
macrophage stimulating 1 receptor (c-met-related tyrosine kinase)
|
|
chr17_-_3407780
|
1.807
|
|
TRPV3
|
transient receptor potential cation channel, subfamily V, member 3
|
|
chr1_+_154390939
|
1.783
|
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A
|
|
chr9_+_94860828
|
1.687
|
|
SUSD3
|
sushi domain containing 3
|
|
chr22_-_28992787
|
1.604
|
NM_020530
|
OSM
|
oncostatin M
|
|
chr14_+_105024576
|
1.482
|
|
CRIP1
|
cysteine-rich protein 1 (intestinal)
|
|
chr17_-_3408038
|
1.450
|
NM_145068
|
TRPV3
|
transient receptor potential cation channel, subfamily V, member 3
|
|
chr9_-_35608300
|
1.399
|
NM_001782
|
CD72
|
CD72 molecule
|
|
chr19_+_12805747
|
1.365
|
|
MAST1
|
microtubule associated serine/threonine kinase 1
|
|
chr9_+_94860769
|
1.343
|
NM_145006
|
SUSD3
|
sushi domain containing 3
|
|
chr8_-_144887876
|
1.296
|
NM_198488
|
FAM83H
|
family with sequence similarity 83, member H
|
|
chr9_-_139068269
|
1.281
|
NM_001246
NM_203468
|
ENTPD2
|
ectonucleoside triphosphate diphosphohydrolase 2
|
|
chr3_+_3816079
|
1.253
|
NM_020873
|
LRRN1
|
leucine rich repeat neuronal 1
|
|
chr17_+_2646481
|
1.240
|
NM_001100398
NM_015085
|
RAP1GAP2
|
RAP1 GTPase activating protein 2
|
|
chr8_+_144888275
|
1.228
|
|
LOC100128338
|
hypothetical LOC100128338
|
|
chr7_-_102044374
|
1.227
|
|
RASA4
|
RAS p21 protein activator 4
|
|
chr1_+_1062137
|
1.216
|
|
LOC254099
|
hypothetical protein LOC254099
|
|
chr19_-_1997229
|
1.213
|
|
MKNK2
|
MAP kinase interacting serine/threonine kinase 2
|
|
chr20_-_61755145
|
1.195
|
NM_015894
|
STMN3
|
stathmin-like 3
|
|
chr17_+_7180571
|
1.174
|
NM_014716
|
ACAP1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1
|
|
chr19_+_1397268
|
1.158
|
|
APC2
|
adenomatosis polyposis coli 2
|
|
chr3_-_49882308
|
1.153
|
NM_024046
|
CAMKV
|
CaM kinase-like vesicle-associated
|
|
chr16_-_4927057
|
1.130
|
NM_002705
|
PPL
|
periplakin
|
|
chr9_+_115367122
|
1.118
|
NM_134427
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr7_+_156496360
|
1.087
|
|
LOC645249
|
hypothetical LOC645249
|
|
chr7_-_156495985
|
1.069
|
NM_005515
|
MNX1
|
motor neuron and pancreas homeobox 1
|
|
chr2_-_85490984
|
1.054
|
|
CAPG
|
capping protein (actin filament), gelsolin-like
|
|
chr11_+_384199
|
1.050
|
NM_007183
|
PKP3
|
plakophilin 3
|
|
chr7_-_102044419
|
1.042
|
NM_001079877
NM_006989
|
RASA4
|
RAS p21 protein activator 4
|
|
chrX_-_48212683
|
1.041
|
|
SLC38A5
|
solute carrier family 38, member 5
|
|
chr15_-_81146188
|
1.040
|
|
AP3B2
|
adaptor-related protein complex 3, beta 2 subunit
|
|
chr19_-_48977247
|
1.017
|
NM_002250
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4
|
|
chr2_+_219453487
|
1.011
|
NM_025216
|
WNT10A
|
wingless-type MMTV integration site family, member 10A
|
|
chr19_+_8180179
|
0.989
|
NM_024552
|
LASS4
|
LAG1 homolog, ceramide synthase 4
|
|
chr12_+_118911859
|
0.976
|
NM_207311
|
CCDC64
|
coiled-coil domain containing 64
|
|
chr1_+_1360765
|
0.964
|
NM_022834
NM_199121
|
VWA1
|
von Willebrand factor A domain containing 1
|
|
chr9_-_99994704
|
0.958
|
NM_052820
|
CORO2A
|
coronin, actin binding protein, 2A
|
|
chr1_-_39929654
|
0.956
|
NM_016257
|
HPCAL4
|
hippocalcin like 4
|
|
chr19_+_1192811
|
0.949
|
|
ATP5D
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit
|
|
chr12_-_120715937
|
0.947
|
NM_019034
|
RHOF
|
ras homolog gene family, member F (in filopodia)
|
|
chr12_+_47658502
|
0.944
|
NM_005430
|
WNT1
|
wingless-type MMTV integration site family, member 1
|
|
chr11_+_384238
|
0.940
|
|
PKP3
|
plakophilin 3
|
|
chr11_+_66927955
|
0.926
|
|
TBC1D10C
|
TBC1 domain family, member 10C
|
|
chr17_+_73654028
|
0.925
|
NM_001163075
|
C17orf99
|
chromosome 17 open reading frame 99
|
|
chr19_-_11311254
|
0.923
|
NM_004283
|
RAB3D
|
RAB3D, member RAS oncogene family
|
|
chr22_-_49317408
|
0.920
|
|
ODF3B
|
outer dense fiber of sperm tails 3B
|
|
chr15_-_19221463
|
0.908
|
|
|
|
|
chr17_-_34015631
|
0.908
|
NM_025248
|
SRCIN1
|
SRC kinase signaling inhibitor 1
|
|
chr12_-_56157898
|
0.896
|
NM_001080156
|
ARHGAP9
|
Rho GTPase activating protein 9
|
|
chr19_-_48976852
|
0.891
|
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4
|
|
chr1_-_164402700
|
0.889
|
|
FAM78B
|
family with sequence similarity 78, member B
|
|
chr19_-_50975121
|
0.885
|
|
|
|
|
chr8_+_145662510
|
0.880
|
NM_145754
|
KIFC2
|
kinesin family member C2
|
|
chr12_+_110328126
|
0.879
|
NM_005475
|
SH2B3
|
SH2B adaptor protein 3
|
|
chr1_+_41057186
|
0.879
|
|
KCNQ4
|
potassium voltage-gated channel, KQT-like subfamily, member 4
|
|
chr11_+_527521
|
0.876
|
NM_198075
|
LRRC56
|
leucine rich repeat containing 56
|
|
chr17_+_8865547
|
0.872
|
NM_004822
|
NTN1
|
netrin 1
|
|
chr12_-_53071285
|
0.870
|
NM_001130967
NM_001130968
|
ZNF385A
|
zinc finger protein 385A
|
|
chr2_+_20729904
|
0.868
|
NM_182828
|
GDF7
|
growth differentiation factor 7
|
|
chr1_-_1041598
|
0.863
|
NM_017891
|
C1orf159
|
chromosome 1 open reading frame 159
|
|
chr11_+_46358909
|
0.845
|
NM_001012334
|
MDK
|
midkine (neurite growth-promoting factor 2)
|
|
chr20_+_36867708
|
0.845
|
NM_001172735
NM_015568
|
PPP1R16B
|
protein phosphatase 1, regulatory (inhibitor) subunit 16B
|
|
chr22_+_23329113
|
0.839
|
NM_001032364
NM_005265
|
GGT1
|
gamma-glutamyltransferase 1
|
|
chr21_+_42512271
|
0.837
|
NM_004915
NM_016818
|
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1
|
|
chr14_+_89597860
|
0.834
|
NM_022054
|
KCNK13
|
potassium channel, subfamily K, member 13
|
|
chr19_-_1463997
|
0.830
|
|
ADAMTSL5
|
ADAMTS-like 5
|
|
chr17_+_54763689
|
0.828
|
NM_001005404
|
YPEL2
|
yippee-like 2 (Drosophila)
|
|
chr19_+_3129735
|
0.821
|
NM_003775
|
S1PR4
|
sphingosine-1-phosphate receptor 4
|
|
chr22_-_22074716
|
0.820
|
|
ZDHHC8P1
|
zinc finger, DHHC-type containing 8 pseudogene 1
|
|
chr6_-_35572694
|
0.819
|
NM_003214
|
TEAD3
|
TEA domain family member 3
|
|
chr16_-_1404635
|
0.813
|
NM_001037125
NM_001193388
|
UNKL
|
unkempt homolog (Drosophila)-like
|
|
chr1_-_37991160
|
0.812
|
|
EPHA10
|
EPH receptor A10
|
|
chr20_-_61639104
|
0.812
|
NM_005975
|
PTK6
|
PTK6 protein tyrosine kinase 6
|
|
chr17_+_7283412
|
0.810
|
NM_004112
|
FGF11
|
fibroblast growth factor 11
|
|
chr2_+_220116986
|
0.804
|
NM_001005209
|
TMEM198
|
transmembrane protein 198
|
|
chr17_-_71352090
|
0.796
|
|
UNC13D
|
unc-13 homolog D (C. elegans)
|
|
chr19_+_45807909
|
0.795
|
|
|
|
|
chr19_-_56841819
|
0.794
|
NM_001098612
|
SIGLEC14
|
sialic acid binding Ig-like lectin 14
|
|
chr2_+_10506233
|
0.794
|
|
|
|
|
chr14_+_30413393
|
0.794
|
NM_001135058
NM_004086
|
COCH
|
coagulation factor C homolog, cochlin (Limulus polyphemus)
|
|
chr19_+_17719504
|
0.782
|
NM_001161357
NM_001161358
|
FCHO1
|
FCH domain only 1
|
|
chr19_-_59666705
|
0.778
|
NM_198988
|
LENG9
|
leukocyte receptor cluster (LRC) member 9
|
|
chr17_-_71352182
|
0.775
|
NM_199242
|
UNC13D
|
unc-13 homolog D (C. elegans)
|
|
chr19_+_1192766
|
0.764
|
|
ATP5D
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit
|
|
chr19_-_1188840
|
0.762
|
NM_152769
|
C19orf26
|
chromosome 19 open reading frame 26
|
|
chr14_-_105245959
|
0.760
|
|
IGHA1
|
immunoglobulin heavy constant alpha 1
|
|
chr15_-_38185865
|
0.759
|
NM_033503
|
BMF
|
Bcl2 modifying factor
|
|
chr8_+_144706699
|
0.754
|
NM_001166237
|
GSDMD
|
gasdermin D
|
|
chr19_+_1192742
|
0.752
|
NM_001001975
NM_001687
|
ATP5D
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit
|
|
chr17_+_7289086
|
0.750
|
NM_000747
|
CHRNB1
|
cholinergic receptor, nicotinic, beta 1 (muscle)
|
|
chr11_+_63864263
|
0.747
|
NM_032251
|
CCDC88B
|
coiled-coil domain containing 88B
|
|
chr20_+_61838479
|
0.747
|
|
LIME1
|
Lck interacting transmembrane adaptor 1
|
|
chr19_+_55614006
|
0.745
|
NM_003121
|
SPIB
|
Spi-B transcription factor (Spi-1/PU.1 related)
|
|
chr16_+_9093035
|
0.743
|
NM_014117
|
C16orf72
|
chromosome 16 open reading frame 72
|
|
chr17_-_71352017
|
0.740
|
|
UNC13D
|
unc-13 homolog D (C. elegans)
|
|
chr19_-_1441340
|
0.735
|
|
PCSK4
|
proprotein convertase subtilisin/kexin type 4
|
|
chr2_+_220117009
|
0.734
|
|
TMEM198
|
transmembrane protein 198
|
|
chr19_+_1192799
|
0.731
|
|
ATP5D
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit
|
|
chr12_-_6615348
|
0.729
|
NM_020400
|
LPAR5
|
lysophosphatidic acid receptor 5
|
|
chr7_+_93976927
|
0.717
|
NM_022900
|
CASD1
|
CAS1 domain containing 1
|
|
chr19_+_868285
|
0.717
|
NM_032551
|
KISS1R
|
KISS1 receptor
|
|
chr3_-_47595190
|
0.716
|
NM_006574
|
CSPG5
|
chondroitin sulfate proteoglycan 5 (neuroglycan C)
|
|
chr5_+_109053628
|
0.713
|
|
MAN2A1
|
mannosidase, alpha, class 2A, member 1
|
|
chr20_+_35964912
|
0.710
|
NM_080607
|
VSTM2L
|
V-set and transmembrane domain containing 2 like
|
|
chr15_+_62230962
|
0.710
|
NM_024798
|
SNX22
|
sorting nexin 22
|
|
chr9_+_135277264
|
0.709
|
|
ADAMTS13
|
ADAM metallopeptidase with thrombospondin type 1 motif, 13
|
|
chr19_+_1192770
|
0.708
|
|
ATP5D
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit
|
|
chr19_+_1192767
|
0.707
|
|
ATP5D
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit
|
|
chr7_-_3049872
|
0.704
|
NM_032415
|
CARD11
|
caspase recruitment domain family, member 11
|
|
chr19_-_1464187
|
0.703
|
NM_213604
|
ADAMTSL5
|
ADAMTS-like 5
|
|
chr11_+_64538161
|
0.701
|
NM_001667
|
ARL2-SNX15
ARL2
|
ARL2-SNX15 read-through transcript
ADP-ribosylation factor-like 2
|
|
chr1_-_159306037
|
0.696
|
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr14_+_93562500
|
0.694
|
|
OTUB2
|
OTU domain, ubiquitin aldehyde binding 2
|
|
chr19_-_13978096
|
0.687
|
NM_002918
|
RFX1
|
regulatory factor X, 1 (influences HLA class II expression)
|
|
chr16_-_87570716
|
0.684
|
NM_005187
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chr16_+_705080
|
0.684
|
NM_024042
|
METRN
|
meteorin, glial cell differentiation regulator
|
|
chr14_-_76564374
|
0.683
|
|
IRF2BPL
|
interferon regulatory factor 2 binding protein-like
|
|
chr16_+_716936
|
0.679
|
NM_207112
NM_032304
|
HAGHL
|
hydroxyacylglutathione hydrolase-like
|
|
chr14_+_104936508
|
0.678
|
|
|
|
|
chr1_-_159306210
|
0.674
|
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr8_-_21827057
|
0.672
|
NM_003974
|
DOK2
|
docking protein 2, 56kDa
|
|
chr19_+_12810258
|
0.672
|
NM_014975
|
MAST1
|
microtubule associated serine/threonine kinase 1
|
|
chr17_-_77478835
|
0.671
|
NM_002359
|
MAFG
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog G (avian)
|
|
chr22_+_23153529
|
0.668
|
NM_000675
|
ADORA2A
|
adenosine A2a receptor
|
|
chr20_-_3696344
|
0.667
|
NM_001039140
|
C20orf27
|
chromosome 20 open reading frame 27
|
|
chr14_-_104333060
|
0.667
|
|
AKT1
|
v-akt murine thymoma viral oncogene homolog 1
|
|
chr11_+_66927976
|
0.662
|
NM_198517
|
TBC1D10C
|
TBC1 domain family, member 10C
|
|
chr16_-_87405817
|
0.660
|
NM_000485
NM_001030018
|
APRT
|
adenine phosphoribosyltransferase
|
|
chrX_-_153252775
|
0.659
|
|
FLNA
|
filamin A, alpha
|
|
chr1_-_181826633
|
0.659
|
NM_001127651
|
NCF2
|
neutrophil cytosolic factor 2
|
|
chr11_-_1549719
|
0.656
|
NM_004420
|
DUSP8
|
dual specificity phosphatase 8
|
|
chr1_-_1346465
|
0.653
|
NM_001145210
|
LOC441869
|
hypothetical protein LOC441869
|
|
chr17_+_72789086
|
0.650
|
NM_001113491
|
SEPT9
|
septin 9
|
|
chr17_+_7289564
|
0.648
|
|
CHRNB1
|
cholinergic receptor, nicotinic, beta 1 (muscle)
|
|
chr20_+_61838389
|
0.648
|
NM_017806
|
LIME1
|
Lck interacting transmembrane adaptor 1
|
|
chr19_+_45728207
|
0.647
|
NM_025213
|
SPTBN4
|
spectrin, beta, non-erythrocytic 4
|
|
chr22_+_48698337
|
0.646
|
|
CRELD2
|
cysteine-rich with EGF-like domains 2
|
|
chr10_+_71060008
|
0.642
|
NM_145306
|
C10orf35
|
chromosome 10 open reading frame 35
|
|
chr5_-_176833259
|
0.641
|
NM_004395
|
DBN1
|
drebrin 1
|
|
chr22_+_18124225
|
0.641
|
NM_005992
NM_080646
NM_080647
|
TBX1
|
T-box 1
|
|
chr1_+_891739
|
0.637
|
NM_001160184
NM_032129
|
PLEKHN1
|
pleckstrin homology domain containing, family N member 1
|
|
chr5_-_131375148
|
0.636
|
NM_001009185
NM_015256
|
ACSL6
|
acyl-CoA synthetase long-chain family member 6
|
|
chr2_-_20288647
|
0.632
|
NM_001006946
|
SDC1
|
syndecan 1
|
|
chr15_+_38437703
|
0.629
|
NM_033510
|
DISP2
|
dispatched homolog 2 (Drosophila)
|
|
chr17_+_75366524
|
0.628
|
NM_005189
NM_032647
|
CBX2
|
chromobox homolog 2
|
|
chr16_+_67698943
|
0.628
|
NM_005329
|
HAS3
|
hyaluronan synthase 3
|
|
chr1_-_159306273
|
0.624
|
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr10_-_103864651
|
0.619
|
NM_003893
|
LDB1
|
LIM domain binding 1
|
|
chr14_-_104333123
|
0.615
|
NM_001014431
NM_001014432
|
AKT1
|
v-akt murine thymoma viral oncogene homolog 1
|
|
chr9_-_138757175
|
0.611
|
NM_001001712
|
LCN10
|
lipocalin 10
|
|
chr19_-_6453224
|
0.611
|
NM_006087
|
TUBB4
|
tubulin, beta 4
|
|
chr21_+_34963557
|
0.608
|
NM_053277
|
CLIC6
|
chloride intracellular channel 6
|
|
chr19_+_10261646
|
0.607
|
NM_003259
|
ICAM5
|
intercellular adhesion molecule 5, telencephalin
|
|
chr11_+_1830763
|
0.602
|
NM_002339
|
LSP1
|
lymphocyte-specific protein 1
|
|
chr6_-_90178685
|
0.598
|
NM_021244
|
RRAGD
|
Ras-related GTP binding D
|
|
chr4_-_83569853
|
0.596
|
|
HNRPDL
|
heterogeneous nuclear ribonucleoprotein D-like
|
|
chr19_+_1200882
|
0.596
|
|
MIDN
|
midnolin
|
|
chr1_-_159306327
|
0.593
|
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr1_+_152014391
|
0.593
|
NM_024330
|
SLC27A3
|
solute carrier family 27 (fatty acid transporter), member 3
|
|
chr3_-_13436725
|
0.591
|
|
NUP210
|
nucleoporin 210kDa
|
|
chr19_-_4017739
|
0.591
|
NM_015898
|
ZBTB7A
|
zinc finger and BTB domain containing 7A
|
|
chr20_+_55399847
|
0.591
|
NM_017495
NM_183425
|
RBM38
|
RNA binding motif protein 38
|
|
chr19_-_19142090
|
0.589
|
NM_001145785
|
MEF2B
|
myocyte enhancer factor 2B
|
|
chr9_-_139184247
|
0.587
|
NM_001013653
|
LRRC26
|
leucine rich repeat containing 26
|
|
chr1_+_158636987
|
0.584
|
NM_020335
|
VANGL2
|
vang-like 2 (van gogh, Drosophila)
|
|
chr20_-_43972982
|
0.584
|
|
PLTP
|
phospholipid transfer protein
|
|
chr17_-_58877276
|
0.583
|
NM_001017916
NM_001915
|
CYB561
|
cytochrome b-561
|
|
chr6_-_31853027
|
0.583
|
NM_025258
|
C6orf27
|
chromosome 6 open reading frame 27
|
|
chr19_+_50535837
|
0.582
|
NM_177417
|
KLC3
|
kinesin light chain 3
|
|
chr19_+_10820105
|
0.581
|
NM_001136482
|
C19orf38
|
chromosome 19 open reading frame 38
|
|
chr6_+_30958672
|
0.580
|
|
DDR1
|
discoidin domain receptor tyrosine kinase 1
|
|
chr9_+_73954174
|
0.577
|
|
GDA
|
guanine deaminase
|
|
chr19_-_50838824
|
0.572
|
NM_001193269
|
EML2
|
echinoderm microtubule associated protein like 2
|
|
chr2_-_26058831
|
0.571
|
|
KIF3C
|
kinesin family member 3C
|
|
chr2_+_232281491
|
0.570
|
|
PTMA
|
prothymosin, alpha
|
|
chr19_-_19142053
|
0.569
|
|
MEF2B
|
myocyte enhancer factor 2B
|
|
chr19_-_53706644
|
0.569
|
|
|
|
|
chr4_-_35922288
|
0.569
|
NM_015230
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2
|
|
chr3_+_198214173
|
0.568
|
|
LOC100507057
MFI2
|
hypothetical LOC100507057
antigen p97 (melanoma associated) identified by monoclonal antibodies 133.2 and 96.5
|
|
chr14_-_56342097
|
0.568
|
NM_172337
|
OTX2
|
orthodenticle homeobox 2
|
|
chr1_-_11663762
|
0.567
|
NM_006341
|
MAD2L2
|
MAD2 mitotic arrest deficient-like 2 (yeast)
|
|
chr4_-_2906256
|
0.565
|
|
MFSD10
|
major facilitator superfamily domain containing 10
|
|
chr1_-_11636983
|
0.563
|
|
FBXO2
|
F-box protein 2
|
|
chr9_+_204867
|
0.563
|
|
DOCK8
|
dedicator of cytokinesis 8
|
|
chr9_+_73954237
|
0.562
|
|
GDA
|
guanine deaminase
|
|
chrX_+_152643516
|
0.562
|
NM_000033
|
ABCD1
|
ATP-binding cassette, sub-family D (ALD), member 1
|
|
chr6_+_2945025
|
0.560
|
NM_000904
|
NQO2
|
NAD(P)H dehydrogenase, quinone 2
|
|
chr10_-_73203202
|
0.560
|
NM_022153
|
C10orf54
|
chromosome 10 open reading frame 54
|
|
chr9_+_204816
|
0.558
|
|
DOCK8
|
dedicator of cytokinesis 8
|
|
chr17_-_73636386
|
0.557
|
NM_001127198
|
TMC6
|
transmembrane channel-like 6
|
|
chr19_+_45728270
|
0.552
|
|
SPTBN4
|
spectrin, beta, non-erythrocytic 4
|
|
chr20_+_34143983
|
0.550
|
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr9_-_139316523
|
0.547
|
NM_001004354
|
NRARP
|
NOTCH-regulated ankyrin repeat protein
|
|
chr22_-_18384251
|
0.546
|
NM_001670
|
ARVCF
|
armadillo repeat gene deleted in velocardiofacial syndrome
|
|
chr19_-_1441403
|
0.545
|
NM_017573
|
PCSK4
|
proprotein convertase subtilisin/kexin type 4
|
|
chr6_+_35373572
|
0.541
|
NM_022047
|
DEF6
|
differentially expressed in FDCP 6 homolog (mouse)
|
|
chr17_-_38150551
|
0.541
|
NM_001991
|
EZH1
|
enhancer of zeste homolog 1 (Drosophila)
|