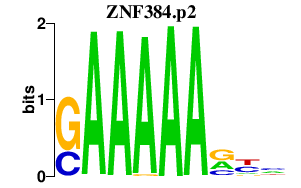
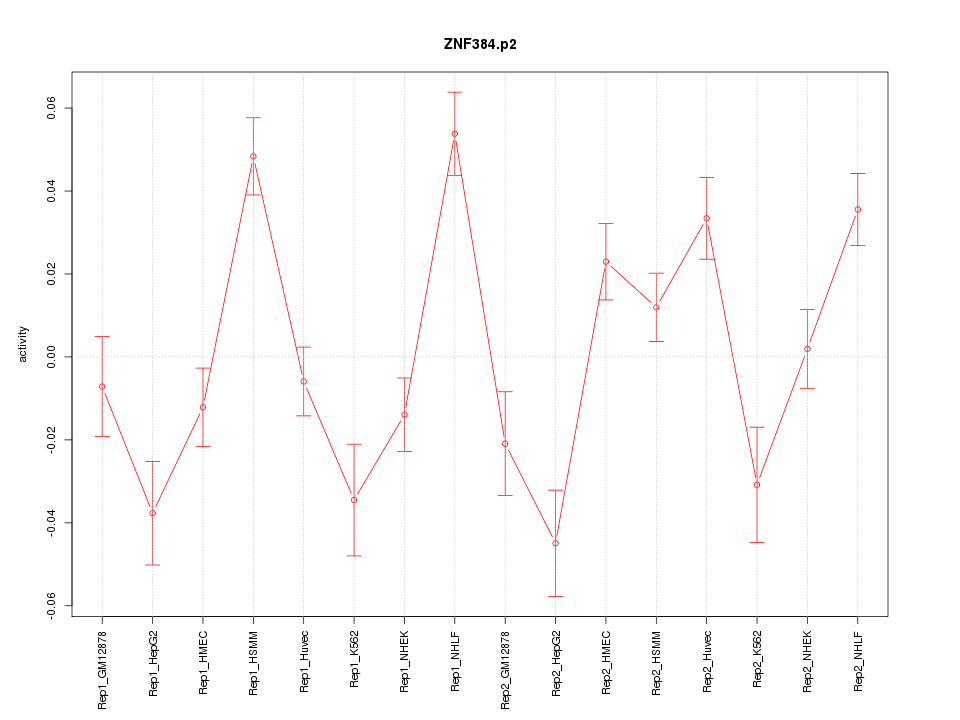
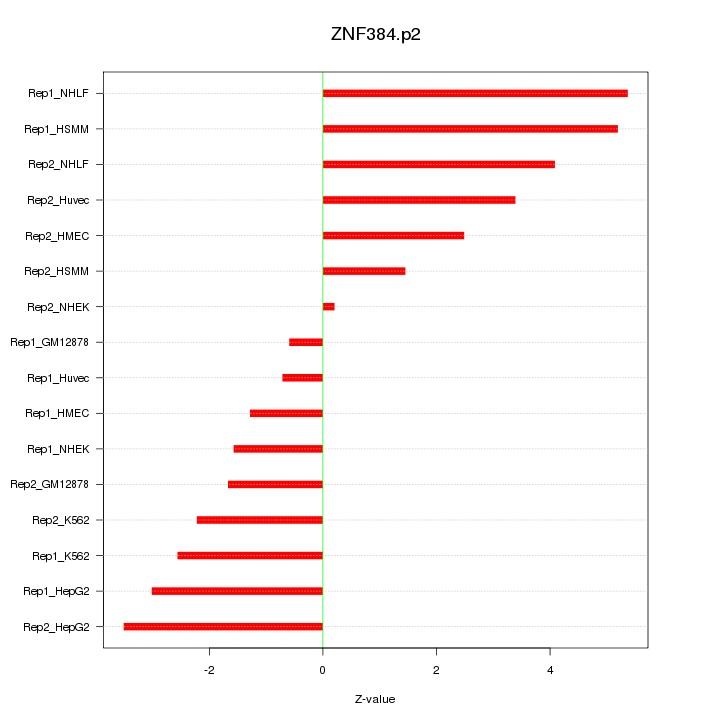
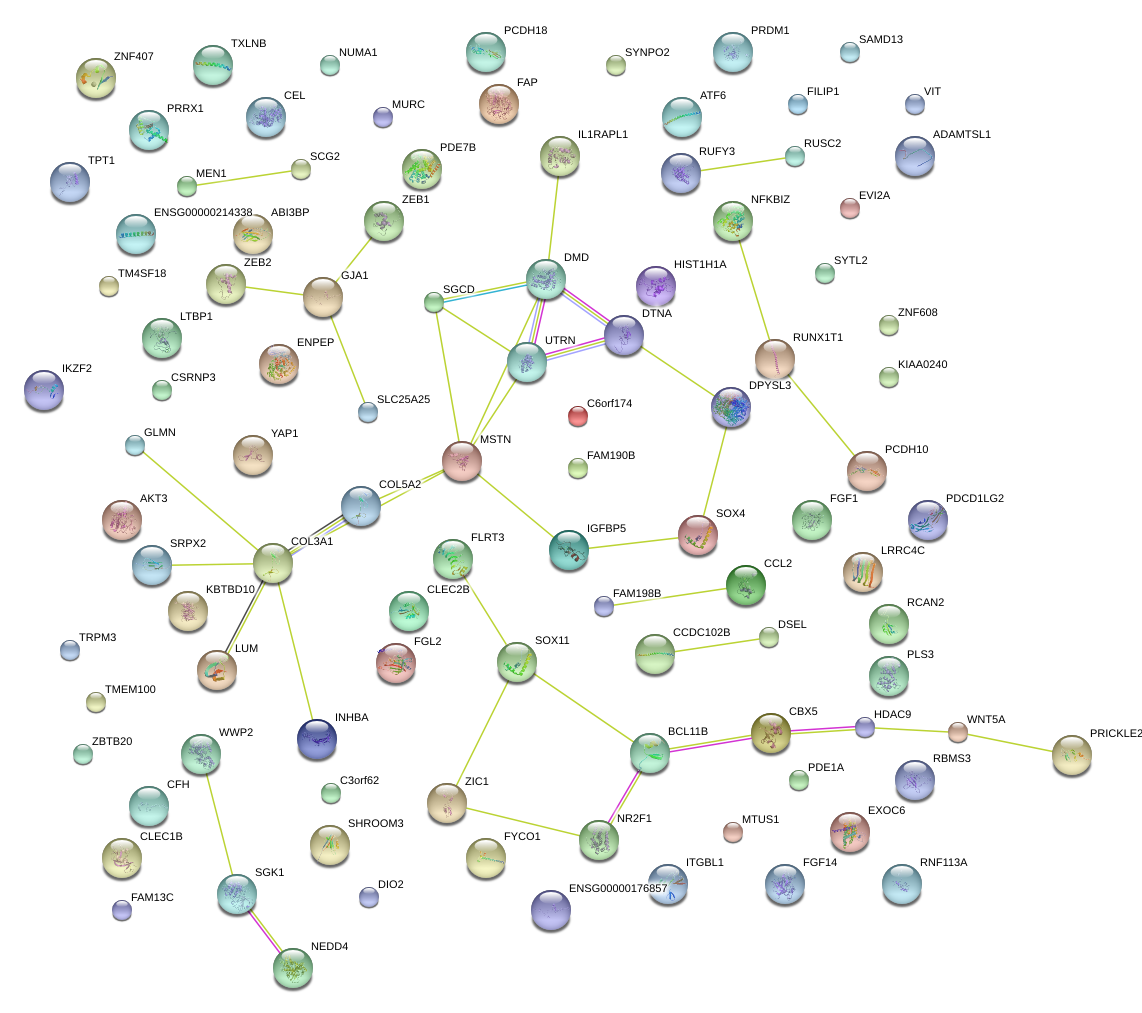
|
chr13_-_101852028
|
6.385
|
NM_175929
|
FGF14
|
fibroblast growth factor 14
|
|
chr1_+_84540921
|
6.308
|
NM_001134664
|
SAMD13
|
sterile alpha motif domain containing 13
|
|
chr5_+_155686750
|
5.802
|
|
SGCD
|
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein)
|
|
chr2_-_189752575
|
5.701
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr11_+_101488380
|
4.969
|
NM_001195045
|
YAP1
|
Yes-associated protein 1
|
|
chr16_+_68516401
|
4.825
|
NM_199424
|
WWP2
|
WW domain containing E3 ubiquitin protein ligase 2
|
|
chr4_+_71806538
|
4.736
|
NM_001037442
NM_014961
|
RUFY3
|
RUN and FYVE domain containing 3
|
|
chr6_+_136214495
|
4.326
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr4_+_134289893
|
4.218
|
NM_020815
NM_032961
|
PCDH10
|
protocadherin 10
|
|
chr1_+_194887630
|
4.159
|
NM_000186
NM_001014975
|
CFH
|
complement factor H
|
|
chr3_-_64186151
|
4.090
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chr5_+_155686333
|
4.062
|
NM_000337
NM_001128209
NM_172244
|
SGCD
|
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein)
|
|
chr17_+_29606408
|
3.976
|
NM_002982
|
CCL2
|
chemokine (C-C motif) ligand 2
|
|
chr6_+_106653429
|
3.968
|
NM_182907
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr6_-_127882192
|
3.955
|
NM_001012279
|
C6orf174
|
chromosome 6 open reading frame 174
|
|
chr2_+_36777336
|
3.932
|
NM_001177969
NM_001177970
NM_001177971
NM_001177972
NM_053276
|
VIT
|
vitrin
|
|
chr11_-_71458739
|
3.900
|
|
NUMA1
|
nuclear mitotic apparatus protein 1
|
|
chr2_+_189547358
|
3.894
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr21_+_25856311
|
3.891
|
|
MIR155HG
|
MIR155 host gene (non-protein coding)
|
|
chr2_-_213723298
|
3.849
|
NM_016260
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr13_+_100902942
|
3.780
|
NM_004791
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains)
|
|
chr2_-_183095308
|
3.707
|
NM_005019
NM_001003683
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent
|
|
chr2_+_189547323
|
3.660
|
NM_000090
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr6_-_76259976
|
3.633
|
NM_015687
|
FILIP1
|
filamin A interacting protein 1
|
|
chr15_-_53996597
|
3.617
|
NM_198400
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr2_-_162808123
|
3.549
|
NM_004460
|
FAP
|
fibroblast activation protein, alpha
|
|
chr7_+_18502342
|
3.545
|
NM_058176
NM_178425
|
HDAC9
|
histone deacetylase 9
|
|
chr2_-_144994349
|
3.537
|
NM_001171653
NM_014795
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr3_+_148609841
|
3.496
|
NM_003412
|
ZIC1
|
Zic family member 1 (odd-paired homolog, Drosophila)
|
|
chr6_+_121798443
|
3.451
|
NM_000165
|
GJA1
|
gap junction protein, alpha 1, 43kDa
|
|
chr2_+_5750229
|
3.428
|
NM_003108
|
SOX11
|
SRY (sex determining region Y)-box 11
|
|
chrX_+_28515479
|
3.231
|
NM_014271
|
IL1RAPL1
|
interleukin 1 receptor accessory protein-like 1
|
|
chr3_-_55489973
|
3.163
|
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr1_-_242073094
|
3.149
|
NM_005465
NM_181690
|
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 (protein kinase B, gamma)
|
|
chr4_+_77575276
|
3.143
|
NM_020859
|
SHROOM3
|
shroom family member 3
|
|
chr9_+_18464138
|
3.125
|
|
ADAMTSL1
|
ADAMTS-like 1
|
|
chr9_-_72673753
|
3.125
|
NM_001007470
NM_020952
NM_024971
NM_206944
NM_206945
NM_206946
NM_206947
NM_206948
|
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3
|
|
chr6_+_42896771
|
3.121
|
NM_015349
|
KIAA0240
|
KIAA0240
|
|
chr3_-_116272911
|
3.085
|
NM_001164343
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr6_-_26125939
|
3.081
|
NM_005325
|
HIST1H1A
|
histone cluster 1, H1a
|
|
chr5_-_146813343
|
2.966
|
NM_001387
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr3_+_148610516
|
2.914
|
|
ZIC1
|
Zic family member 1 (odd-paired homolog, Drosophila)
|
|
chr2_+_189547377
|
2.904
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr9_+_35528890
|
2.870
|
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr2_+_33213111
|
2.856
|
NM_000627
NM_001166264
NM_001166265
NM_001166266
|
LTBP1
|
latent transforming growth factor beta binding protein 1
|
|
chr2_-_144994038
|
2.847
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr3_+_103029523
|
2.726
|
NM_001005474
|
NFKBIZ
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta
|
|
chr4_+_120168002
|
2.722
|
|
SYNPO2
|
synaptopodin 2
|
|
chr11_-_85115105
|
2.720
|
NM_206927
NM_206928
|
SYTL2
|
synaptotagmin-like 2
|
|
chr5_-_141981083
|
2.698
|
NM_001144892
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr18_+_70471906
|
2.654
|
NM_001146189
NM_001146190
NM_017757
|
ZNF407
|
zinc finger protein 407
|
|
chr3_-_150533948
|
2.648
|
NM_001184723
NM_138786
|
TM4SF18
|
transmembrane 4 L six family member 18
|
|
chr18_+_30427279
|
2.627
|
NM_001128175
NM_001198938
NM_001198939
NM_001198940
|
DTNA
|
dystrobrevin, alpha
|
|
chr4_+_111616630
|
2.585
|
NM_001977
|
ENPEP
|
glutamyl aminopeptidase (aminopeptidase A)
|
|
chr10_+_86174665
|
2.560
|
|
FAM190B
|
family with sequence similarity 190, member B
|
|
chrX_+_114734074
|
2.550
|
NM_001136025
NM_001172335
|
PLS3
|
plastin 3
|
|
chr9_+_102380156
|
2.548
|
NM_001018116
|
MURC
|
muscle-related coiled-coil protein
|
|
chr12_-_52939636
|
2.547
|
NM_001127321
|
CBX5
|
chromobox homolog 5
|
|
chr8_-_93181091
|
2.527
|
NM_001198628
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr6_-_46401174
|
2.521
|
|
RCAN2
|
regulator of calcineurin 2
|
|
chr7_-_41709191
|
2.466
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chr14_-_98807307
|
2.432
|
|
BCL11B
|
B-cell CLL/lymphoma 11B (zinc finger protein)
|
|
chr3_-_49289285
|
2.402
|
NM_198562
|
C3orf62
|
chromosome 3 open reading frame 62
|
|
chr12_-_52939584
|
2.398
|
NM_001127322
|
CBX5
|
chromobox homolog 5
|
|
chr10_-_60792225
|
2.387
|
NM_001001971
NM_001166698
NM_198215
|
FAM13C
|
family with sequence similarity 13, member C
|
|
chr17_-_51155073
|
2.366
|
NM_018286
|
TMEM100
|
transmembrane protein 100
|
|
chr9_+_35528628
|
2.356
|
NM_001135999
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr17_-_26672757
|
2.343
|
NM_001003927
NM_014210
|
EVI2A
|
ecotropic viral integration site 2A
|
|
chr10_+_94584449
|
2.323
|
NM_001013848
|
EXOC6
|
exocyst complex component 6
|
|
chr18_+_30544193
|
2.314
|
NM_001198941
NM_032978
|
DTNA
|
dystrobrevin, alpha
|
|
chr9_+_129893948
|
2.305
|
NM_001006642
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr8_-_17599438
|
2.292
|
NM_020749
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr3_-_102194938
|
2.259
|
NM_015429
|
ABI3BP
|
ABI family, member 3 (NESH) binding protein
|
|
chr3_+_66527586
|
2.245
|
|
|
|
|
chr13_-_44812246
|
2.241
|
|
TPT1
|
tumor protein, translationally-controlled 1
|
|
chr18_+_64616296
|
2.241
|
NM_024781
|
CCDC102B
|
coiled-coil domain containing 102B
|
|
chr4_-_138673014
|
2.237
|
|
PCDH18
|
protocadherin 18
|
|
chr12_-_90029672
|
2.230
|
NM_002345
|
LUM
|
lumican
|
|
chr11_-_40271086
|
2.224
|
|
LRRC4C
|
leucine rich repeat containing 4C
|
|
chrX_-_33139349
|
2.218
|
NM_004006
|
DMD
|
dystrophin
|
|
chrX_+_99785818
|
2.196
|
NM_014467
|
SRPX2
|
sushi-repeat containing protein, X-linked 2
|
|
chr2_-_190635566
|
2.193
|
NM_005259
|
MSTN
|
myostatin
|
|
chr2_-_217268492
|
2.179
|
NM_000599
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr3_+_29297806
|
2.152
|
NM_001003792
NM_001003793
NM_001177711
NM_001177712
NM_014483
|
RBMS3
|
RNA binding motif, single stranded interacting protein 3
|
|
chr14_-_79747592
|
2.148
|
NM_001007023
|
DIO2
|
deiodinase, iodothyronine, type II
|
|
chr2_-_224175386
|
2.146
|
NM_003469
|
SCG2
|
secretogranin II
|
|
chr9_+_5500544
|
2.124
|
NM_025239
|
PDCD1LG2
|
programmed cell death 1 ligand 2
|
|
chr6_-_134680888
|
2.108
|
NM_001143676
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr1_+_168898898
|
2.107
|
|
PRRX1
|
paired related homeobox 1
|
|
chr2_+_170074457
|
2.090
|
NM_006063
|
KBTBD10
|
kelch repeat and BTB (POZ) domain containing 10
|
|
chr2_+_166034402
|
2.080
|
NM_001172173
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3
|
|
chr18_-_63333487
|
2.061
|
|
DSEL
|
dermatan sulfate epimerase-like
|
|
chr2_+_36777406
|
2.060
|
|
VIT
|
vitrin
|
|
chr7_-_76667045
|
2.036
|
NM_006682
|
FGL2
|
fibrinogen-like 2
|
|
chr20_-_14266200
|
2.024
|
NM_013281
NM_198391
|
FLRT3
|
fibronectin leucine rich transmembrane protein 3
|
|
chr5_+_92944680
|
2.015
|
NM_005654
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr4_-_159313167
|
2.007
|
NM_016613
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr4_-_138673078
|
2.005
|
NM_019035
|
PCDH18
|
protocadherin 18
|
|
chr5_-_124108672
|
1.995
|
NM_020747
|
ZNF608
|
zinc finger protein 608
|
|
chr1_+_160002733
|
1.987
|
|
ATF6
|
activating transcription factor 6
|
|
chr12_-_9913724
|
1.982
|
NM_005127
|
CLEC2B
|
C-type lectin domain family 2, member B
|
|
chr6_-_139654807
|
1.979
|
NM_153235
|
TXLNB
|
taxilin beta
|
|
chr1_-_103346639
|
1.978
|
NM_001190709
NM_001854
NM_080629
NM_080630
|
COL11A1
|
collagen, type XI, alpha 1
|
|
chr4_-_186934059
|
1.969
|
NM_001145673
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr1_-_42156957
|
1.953
|
|
HIVEP3
|
human immunodeficiency virus type I enhancer binding protein 3
|
|
chr4_+_159311353
|
1.950
|
|
LOC285505
|
hypothetical protein LOC285505
|
|
chr5_+_125786975
|
1.919
|
|
GRAMD3
|
GRAM domain containing 3
|
|
chr15_-_78050515
|
1.917
|
NM_001114735
NM_004049
|
BCL2A1
|
BCL2-related protein A1
|
|
chr8_-_13018123
|
1.915
|
NM_001164271
|
DLC1
|
deleted in liver cancer 1
|
|
chr1_+_160002693
|
1.899
|
NM_007348
|
ATF6
|
activating transcription factor 6
|
|
chr2_-_188127430
|
1.881
|
NM_001032281
NM_006287
|
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor)
|
|
chr8_-_23768242
|
1.876
|
NM_003155
|
STC1
|
stanniocalcin 1
|
|
chr6_+_74462654
|
1.871
|
|
CD109
|
CD109 molecule
|
|
chr6_+_12398514
|
1.870
|
NM_001168319
NM_001955
|
EDN1
|
endothelin 1
|
|
chr2_-_231697999
|
1.858
|
NM_000867
|
HTR2B
|
5-hydroxytryptamine (serotonin) receptor 2B
|
|
chr1_+_100957883
|
1.854
|
NM_001078
NM_080682
|
VCAM1
|
vascular cell adhesion molecule 1
|
|
chr1_+_46151845
|
1.848
|
|
MAST2
|
microtubule associated serine/threonine kinase 2
|
|
chr9_-_16717887
|
1.824
|
|
BNC2
|
basonuclin 2
|
|
chr21_-_27261276
|
1.818
|
NM_007038
|
ADAMTS5
|
ADAM metallopeptidase with thrombospondin type 1 motif, 5
|
|
chr1_-_101132833
|
1.810
|
|
EXTL2
|
exostoses (multiple)-like 2
|
|
chr9_-_36266930
|
1.792
|
NM_001128227
NM_001190388
|
GNE
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase
|
|
chrX_+_100361588
|
1.791
|
NM_001171184
NM_001939
|
DRP2
|
dystrophin related protein 2
|
|
chr3_+_58459131
|
1.779
|
NM_153331
|
KCTD6
|
potassium channel tetramerisation domain containing 6
|
|
chr12_+_79625538
|
1.771
|
NM_002469
|
MYF6
|
myogenic factor 6 (herculin)
|
|
chr7_-_27108826
|
1.765
|
NM_006735
|
HOXA2
|
homeobox A2
|
|
chr12_+_13240868
|
1.752
|
NM_001423
|
EMP1
|
epithelial membrane protein 1
|
|
chr1_-_42156913
|
1.732
|
|
|
|
|
chr2_-_198248828
|
1.726
|
NM_144629
|
RFTN2
|
raftlin family member 2
|
|
chr1_+_185064654
|
1.724
|
NM_024420
|
PLA2G4A
|
phospholipase A2, group IVA (cytosolic, calcium-dependent)
|
|
chr7_+_93388951
|
1.715
|
NM_004126
|
GNG11
|
guanine nucleotide binding protein (G protein), gamma 11
|
|
chr3_+_45042754
|
1.694
|
NM_003278
|
CLEC3B
|
C-type lectin domain family 3, member B
|
|
chr17_-_36436979
|
1.690
|
NM_031957
|
KRTAP1-5
|
keratin associated protein 1-5
|
|
chr7_+_93862137
|
1.686
|
|
COL1A2
PLAG1
|
collagen, type I, alpha 2
pleiomorphic adenoma gene 1
|
|
chr8_-_6407882
|
1.683
|
NM_001118887
NM_001118888
NM_001147
|
ANGPT2
|
angiopoietin 2
|
|
chr10_-_98315090
|
1.680
|
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr1_+_242281199
|
1.654
|
|
ZNF238
|
zinc finger protein 238
|
|
chr1_-_23393808
|
1.652
|
NM_000864
|
HTR1D
|
5-hydroxytryptamine (serotonin) receptor 1D
|
|
chr5_-_138238877
|
1.646
|
|
LRRTM2
|
leucine rich repeat transmembrane neuronal 2
|
|
chr8_-_72918697
|
1.634
|
|
MSC
|
musculin
|
|
chr12_-_90100733
|
1.632
|
NM_001920
|
DCN
|
decorin
|
|
chr18_-_51406841
|
1.624
|
NM_001083962
NM_003199
|
TCF4
|
transcription factor 4
|
|
chr4_-_147079018
|
1.623
|
NM_178835
|
ZNF827
|
zinc finger protein 827
|
|
chr17_-_44047299
|
1.615
|
NM_024016
|
HOXB8
|
homeobox B8
|
|
chrX_-_135691168
|
1.611
|
NM_004840
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr4_-_147079410
|
1.602
|
|
|
|
|
chr14_+_95792299
|
1.597
|
NM_000710
|
BDKRB1
|
bradykinin receptor B1
|
|
chr17_-_36469869
|
1.597
|
NM_001165252
|
LOC730755
|
keratin associated protein 2-4-like
|
|
chr1_+_160002746
|
1.596
|
|
ATF6
|
activating transcription factor 6
|
|
chr17_-_44006808
|
1.590
|
NM_002146
|
HOXB3
|
homeobox B3
|
|
chr1_-_42157077
|
1.579
|
NM_001127714
NM_024503
|
HIVEP3
|
human immunodeficiency virus type I enhancer binding protein 3
|
|
chr13_-_98428244
|
1.573
|
NM_001130048
NM_001130050
|
DOCK9
|
dedicator of cytokinesis 9
|
|
chr10_+_88720477
|
1.567
|
NM_133447
|
AGAP11
|
ankyrin repeat and GTPase domain Arf GTPase activating protein 11
|
|
chr12_+_74160779
|
1.561
|
NM_006851
|
GLIPR1
|
GLI pathogenesis-related 1
|
|
chr2_-_133144094
|
1.561
|
|
LYPD1
|
LY6/PLAUR domain containing 1
|
|
chr3_+_88270951
|
1.550
|
NM_018293
|
ZNF654
|
zinc finger protein 654
|
|
chr21_+_44002290
|
1.540
|
|
PDXK
|
pyridoxal (pyridoxine, vitamin B6) kinase
|
|
chr21_-_38955487
|
1.539
|
NM_001136154
NM_004449
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr8_-_72919161
|
1.536
|
|
MSC
|
musculin
|
|
chr15_+_65217412
|
1.533
|
NM_001145103
|
SMAD3
|
SMAD family member 3
|
|
chr5_+_178419938
|
1.530
|
NM_014594
|
ZNF354C
|
zinc finger protein 354C
|
|
chr6_+_152053323
|
1.523
|
NM_001122742
|
ESR1
|
estrogen receptor 1
|
|
chr12_+_79634820
|
1.520
|
NM_005593
|
MYF5
|
myogenic factor 5
|
|
chr5_-_138238891
|
1.519
|
|
CTNNA1
LRRTM2
|
catenin (cadherin-associated protein), alpha 1, 102kDa
leucine rich repeat transmembrane neuronal 2
|
|
chr6_-_166321393
|
1.515
|
|
C6orf176
|
chromosome 6 open reading frame 176
|
|
chr1_-_103346482
|
1.514
|
|
COL11A1
|
collagen, type XI, alpha 1
|
|
chr17_-_24491521
|
1.510
|
|
MYO18A
|
myosin XVIIIA
|
|
chr1_+_169747788
|
1.493
|
|
PRRC2C
|
proline-rich coiled-coil 2C
|
|
chr5_-_58607673
|
1.493
|
NM_001197220
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr16_+_3295433
|
1.488
|
NM_153028
|
ZNF75A
|
zinc finger protein 75a
|
|
chr3_-_167037861
|
1.488
|
|
BCHE
|
butyrylcholinesterase
|
|
chr12_+_52733927
|
1.486
|
NM_153633
|
HOXC4
HOXC6
|
homeobox C4
homeobox C6
|
|
chr4_+_54790020
|
1.485
|
NM_006206
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr12_-_2814410
|
1.482
|
NM_031474
|
NRIP2
|
nuclear receptor interacting protein 2
|
|
chr12_-_48902692
|
1.472
|
NM_001113547
|
LIMA1
|
LIM domain and actin binding 1
|
|
chr20_+_19818209
|
1.469
|
NM_018993
|
RIN2
|
Ras and Rab interactor 2
|
|
chr6_+_106640903
|
1.466
|
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr4_-_152368492
|
1.465
|
|
SH3D19
|
SH3 domain containing 19
|
|
chr13_+_49487390
|
1.458
|
NM_173605
NM_199464
|
TRIM13
KCNRG
|
tripartite motif containing 13
potassium channel regulator
|
|
chr14_+_62740878
|
1.456
|
NM_020663
|
RHOJ
|
ras homolog gene family, member J
|
|
chr8_+_72918772
|
1.454
|
|
LOC100132891
|
hypothetical LOC100132891
|
|
chr1_+_162795328
|
1.451
|
NM_002585
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr4_-_159312973
|
1.448
|
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr1_-_171442937
|
1.440
|
NM_003326
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4
|
|
chr9_+_102231616
|
1.437
|
NM_001198806
|
C9orf30
|
chromosome 9 open reading frame 30
|
|
chr8_+_72918889
|
1.437
|
|
|
|
|
chr1_+_66772393
|
1.431
|
NM_032291
|
SGIP1
|
SH3-domain GRB2-like (endophilin) interacting protein 1
|
|
chr1_+_200775864
|
1.430
|
NM_001197131
|
PPP1R12B
|
protein phosphatase 1, regulatory (inhibitor) subunit 12B
|
|
chr11_-_56846246
|
1.423
|
|
TNKS1BP1
|
tankyrase 1 binding protein 1, 182kDa
|
|
chr2_+_170742900
|
1.421
|
NM_001083615
NM_001171642
NM_138995
|
MYO3B
|
myosin IIIB
|
|
chr2_+_151922350
|
1.414
|
NM_007115
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6
|
|
chr5_-_138238953
|
1.409
|
NM_015564
|
LRRTM2
|
leucine rich repeat transmembrane neuronal 2
|
|
chr1_-_79244948
|
1.404
|
NM_022159
|
ELTD1
|
EGF, latrophilin and seven transmembrane domain containing 1
|
|
chr7_+_17349425
|
1.397
|
|
AHR
|
aryl hydrocarbon receptor
|
|
chr8_-_72918938
|
1.396
|
|
MSC
|
musculin
|
|
chr8_-_120720200
|
1.394
|
NM_001040092
NM_001130863
NM_006209
|
ENPP2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2
|
|
chr2_+_202379442
|
1.393
|
NM_139158
|
CDK15
|
cyclin-dependent kinase 15
|
|
chr1_-_197173159
|
1.392
|
|
LOC100131234
|
familial acute myelogenous leukemia related factor
|
|
chr12_-_9913199
|
1.387
|
|
CLEC2B
|
C-type lectin domain family 2, member B
|
|
chr1_+_160868822
|
1.384
|
NM_001014796
NM_006182
|
DDR2
|
discoidin domain receptor tyrosine kinase 2
|
|
chr2_-_165520241
|
1.375
|
NM_173512
|
SLC38A11
|
solute carrier family 38, member 11
|