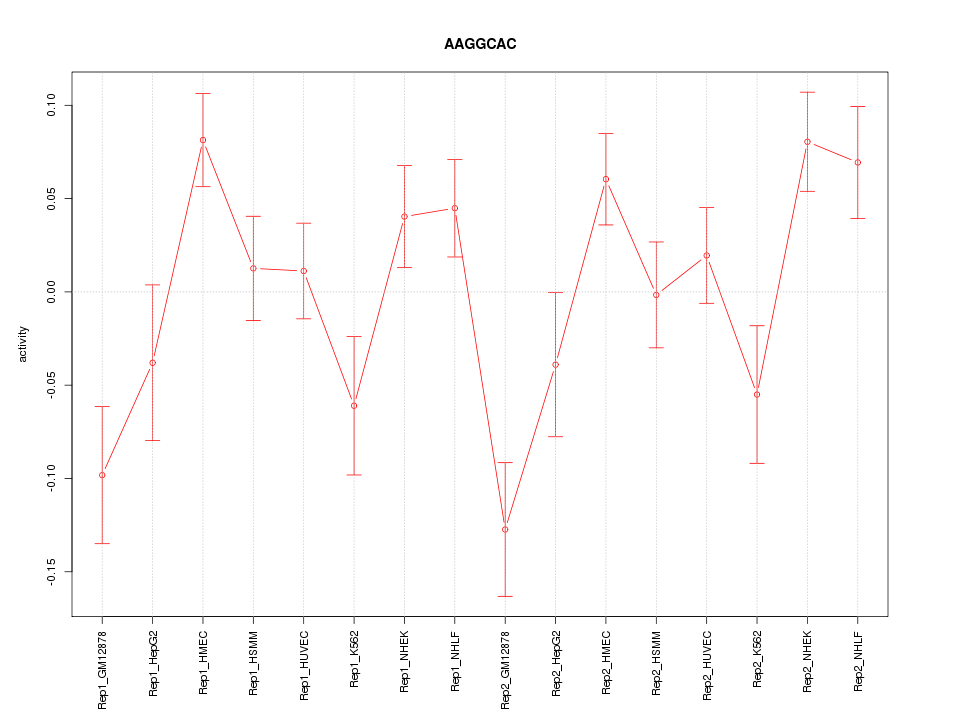
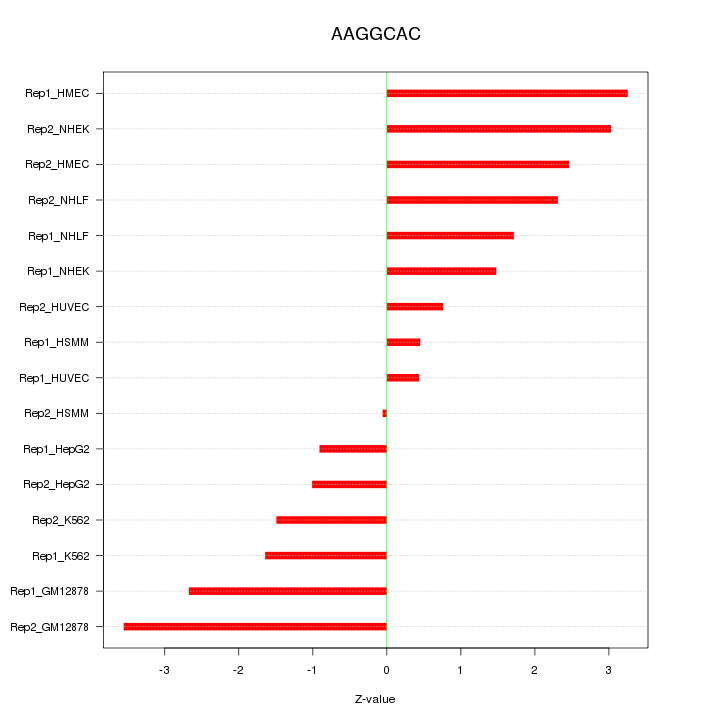
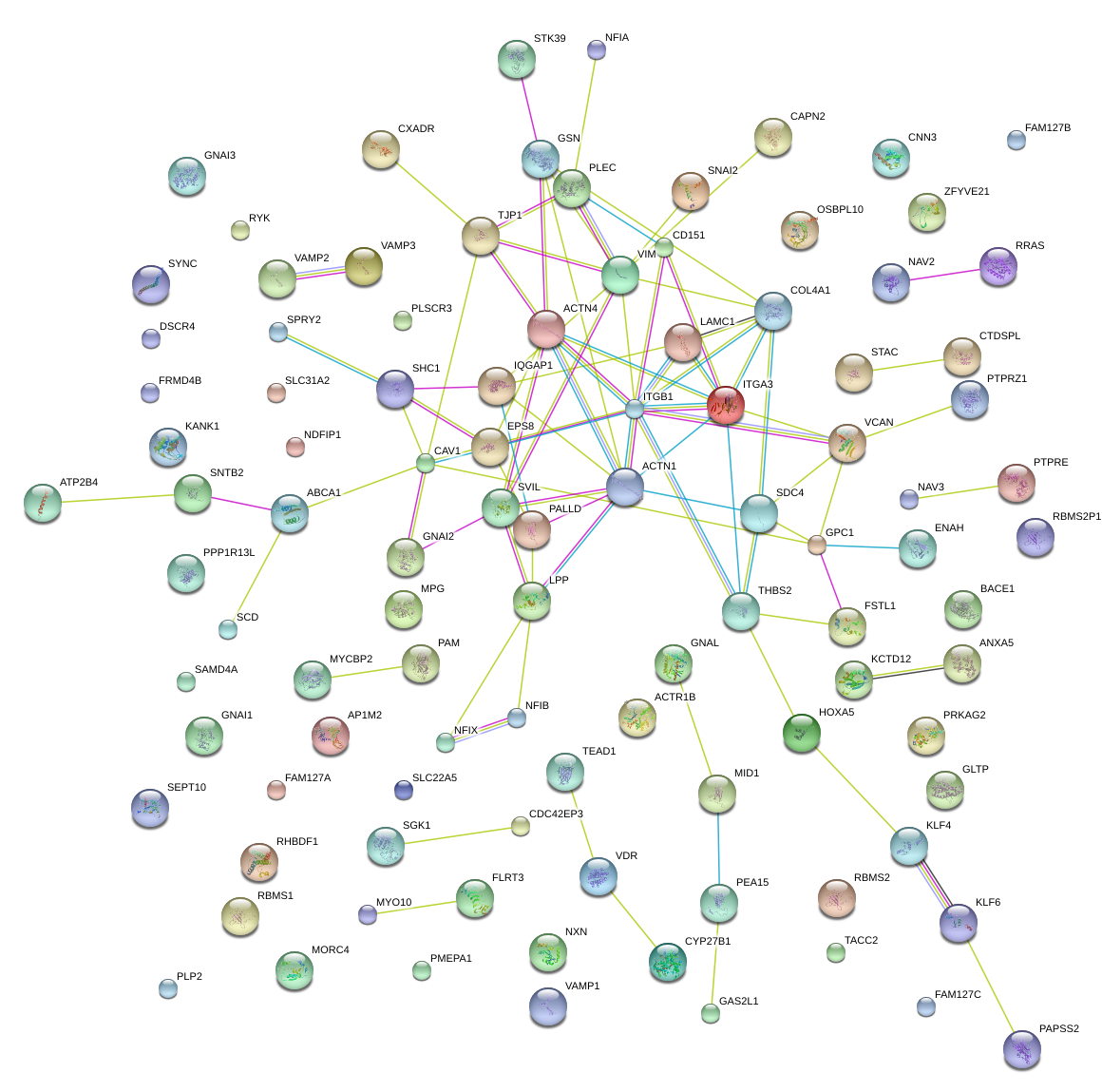
|
chr7_+_116166346
|
5.521
|
NM_001172896
NM_001172897
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chr7_+_116165043
|
5.308
|
NM_001172895
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chr7_+_116164838
|
5.294
|
NM_001753
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chr8_-_49833823
|
4.220
|
NM_003068
|
SNAI2
|
snail homolog 2 (Drosophila)
|
|
chr3_-_120169493
|
4.113
|
NM_007085
|
FSTL1
|
follistatin-like 1
|
|
chr4_+_169418167
|
4.113
|
NM_001166108
NM_016081
|
PALLD
|
palladin, cytoskeletal associated protein
|
|
chr19_-_50143350
|
3.914
|
NM_006270
|
RRAS
|
related RAS viral (r-ras) oncogene homolog
|
|
chr4_+_169753105
|
3.881
|
NM_001166110
|
PALLD
|
palladin, cytoskeletal associated protein
|
|
chr4_+_169552752
|
3.865
|
NM_001166109
|
PALLD
|
palladin, cytoskeletal associated protein
|
|
chr17_-_767287
|
3.772
|
NM_001205319
|
NXN
|
nucleoredoxin
|
|
chr17_-_882905
|
3.583
|
NM_022463
|
NXN
|
nucleoredoxin
|
|
chr13_-_77460432
|
3.259
|
NM_138444
|
KCTD12
|
potassium channel tetramerisation domain containing 12
|
|
chr2_-_161350304
|
3.206
|
NM_002897
NM_016836
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr9_+_115913227
|
3.192
|
NM_001860
|
SLC31A2
|
solute carrier family 31 (copper transporters), member 2
|
|
chr1_-_225840660
|
3.162
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr20_-_14318200
|
3.068
|
NM_013281
NM_198391
|
FLRT3
|
fibronectin leucine rich transmembrane protein 3
|
|
chr17_+_48133331
|
2.953
|
NM_002204
NM_005501
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor)
|
|
chr3_-_32023237
|
2.913
|
NM_001174060
NM_017784
|
OSBPL10
|
oxysterol binding protein-like 10
|
|
chr5_-_16936371
|
2.861
|
NM_012334
|
MYO10
|
myosin X
|
|
chr1_+_223900061
|
2.847
|
NM_001748
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr9_+_706744
|
2.823
|
NM_153186
|
KANK1
|
KN motif and ankyrin repeat domains 1
|
|
chr6_-_169654047
|
2.816
|
NM_003247
|
THBS2
|
thrombospondin 2
|
|
chr7_+_79764087
|
2.774
|
NM_002069
|
GNAI1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1
|
|
chr9_+_504661
|
2.688
|
NM_015158
|
KANK1
|
KN motif and ankyrin repeat domains 1
|
|
chr13_-_110959441
|
2.592
|
NM_001845
|
COL4A1
|
collagen, type IV, alpha 1
|
|
chr6_-_134498973
|
2.556
|
NM_001143677
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chrX_-_10588458
|
2.542
|
NM_000381
NM_033289
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr6_-_134639195
|
2.503
|
NM_001143676
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr1_-_95392501
|
2.470
|
NM_001839
|
CNN3
|
calponin 3, acidic
|
|
chrX_-_10544852
|
2.405
|
NM_001193277
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chrX_-_10645726
|
2.403
|
NM_001098624
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr6_-_134496006
|
2.385
|
NM_005627
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr1_+_223889253
|
2.365
|
NM_001146068
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr7_-_151574209
|
2.325
|
NM_016203
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit
|
|
chr7_-_27183225
|
2.318
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chr20_-_43977023
|
2.300
|
NM_002999
|
SDC4
|
syndecan 4
|
|
chr10_+_17270257
|
2.295
|
NM_003380
|
VIM
|
vimentin
|
|
chr1_-_154946859
|
2.266
|
NM_001130041
NM_001202859
NM_003029
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1
|
|
chrX_-_10851808
|
2.240
|
NM_033290
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr9_-_14398981
|
2.237
|
NM_001190738
|
NFIB
|
nuclear factor I/B
|
|
chr7_-_151511925
|
2.159
|
NM_001040633
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit
|
|
chr6_-_134497069
|
2.151
|
NM_001143678
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr5_+_102201526
|
2.143
|
NM_000919
NM_001177306
NM_138766
NM_138821
NM_138822
|
PAM
|
peptidylglycine alpha-amidating monooxygenase
|
|
chrX_+_134166318
|
2.123
|
NM_001078171
|
FAM127A
|
family with sequence similarity 127, member A
|
|
chr7_-_151329153
|
2.111
|
NM_024429
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit
|
|
chr19_-_45909566
|
2.104
|
NM_001142502
|
PPP1R13L
|
protein phosphatase 1, regulatory subunit 13 like
|
|
chr12_+_78225068
|
2.071
|
NM_014903
|
NAV3
|
neuron navigator 3
|
|
chr1_+_7831326
|
2.050
|
NM_004781
|
VAMP3
|
vesicle-associated membrane protein 3 (cellubrevin)
|
|
chr20_-_56286540
|
2.050
|
NM_199171
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1
|
|
chr9_+_124061880
|
2.032
|
NM_000177
|
GSN
|
gelsolin
|
|
chr12_-_15942350
|
1.994
|
NM_004447
|
EPS8
|
epidermal growth factor receptor pathway substrate 8
|
|
chr10_+_89419352
|
1.990
|
NM_001015880
NM_004670
|
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2
|
|
chr3_+_187943192
|
1.944
|
NM_001167671
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr9_-_14314036
|
1.939
|
NM_001190737
NM_005596
|
NFIB
|
nuclear factor I/B
|
|
chr19_-_45908288
|
1.914
|
NM_006663
|
PPP1R13L
|
protein phosphatase 1, regulatory subunit 13 like
|
|
chr1_+_203595897
|
1.908
|
NM_001001396
NM_001684
|
ATP2B4
|
ATPase, Ca++ transporting, plasma membrane 4
|
|
chr2_-_169103886
|
1.898
|
NM_013233
|
STK39
|
serine threonine kinase 39
|
|
chr10_-_33247178
|
1.892
|
NM_002211
|
ITGB1
|
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12)
|
|
chr7_+_121513158
|
1.891
|
NM_001206838
NM_001206839
NM_002851
|
PTPRZ1
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1
|
|
chr11_+_832909
|
1.872
|
NM_001039490
NM_004357
NM_139029
NM_139030
|
CD151
|
CD151 molecule (Raph blood group)
|
|
chr10_-_30024594
|
1.855
|
NM_003174
|
SVIL
|
supervillin
|
|
chr16_+_69220940
|
1.844
|
NM_006750
|
SNTB2
|
syntrophin, beta 2 (dystrophin-associated protein A1, 59kDa, basic component 2)
|
|
chrX_+_49028183
|
1.836
|
NM_002668
|
PLP2
|
proteolipid protein 2 (colonic epithelium-enriched)
|
|
chr3_+_187871473
|
1.819
|
NM_001167672
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr16_-_122573
|
1.801
|
NM_022450
|
RHBDF1
|
rhomboid 5 homolog 1 (Drosophila)
|
|
chr10_-_33246786
|
1.794
|
NM_133376
|
ITGB1
|
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12)
|
|
chr3_-_69435429
|
1.788
|
NM_015123
|
FRMD4B
|
FERM domain containing 4B
|
|
chr12_-_48298765
|
1.788
|
NM_000376
NM_001017535
NM_001017536
|
VDR
|
vitamin D (1,25- dihydroxyvitamin D3) receptor
|
|
chr11_+_12695851
|
1.740
|
NM_021961
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor)
|
|
chr8_-_145013628
|
1.717
|
NM_201384
|
PLEC
|
plectin
|
|
chr12_-_110318178
|
1.711
|
NM_016433
|
GLTP
|
glycolipid transfer protein
|
|
chr8_-_145025043
|
1.711
|
NM_201380
|
PLEC
|
plectin
|
|
chr20_-_56285576
|
1.710
|
NM_199169
NM_199170
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1
|
|
chr20_-_56284945
|
1.695
|
NM_020182
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1
|
|
chr8_-_145027975
|
1.685
|
NM_201379
|
PLEC
|
plectin
|
|
chr3_+_187930720
|
1.666
|
NM_005578
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr15_-_30114536
|
1.649
|
NM_003257
NM_175610
|
TJP1
|
tight junction protein 1 (zona occludens 1)
|
|
chr8_-_145047639
|
1.646
|
NM_201378
|
PLEC
|
plectin
|
|
chr1_+_182992329
|
1.625
|
NM_002293
|
LAMC1
|
laminin, gamma 1 (formerly LAMB2)
|
|
chr8_-_145018057
|
1.624
|
NM_201382
|
PLEC
|
plectin
|
|
chrX_-_106243442
|
1.581
|
NM_001085354
NM_024657
|
MORC4
|
MORC family CW-type zinc finger 4
|
|
chr1_-_154943001
|
1.574
|
NM_001130040
NM_183001
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1
|
|
chr8_-_145016691
|
1.571
|
NM_201383
|
PLEC
|
plectin
|
|
chr8_-_145018904
|
1.567
|
NM_201381
|
PLEC
|
plectin
|
|
chr9_-_107690526
|
1.549
|
NM_005502
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr10_+_129705299
|
1.545
|
NM_006504
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E
|
|
chr19_+_39138265
|
1.527
|
NM_004924
|
ACTN4
|
actinin, alpha 4
|
|
chr21_+_18885104
|
1.522
|
NM_001207063
NM_001207064
NM_001207065
NM_001207066
NM_001338
|
CXADR
|
coxsackie virus and adenovirus receptor
|
|
chr9_+_124030370
|
1.515
|
NM_001127662
NM_001127663
NM_001127664
NM_001127665
NM_001127666
NM_001127667
NM_198252
|
GSN
|
gelsolin
|
|
chr9_-_110252045
|
1.503
|
NM_004235
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr10_+_129845806
|
1.501
|
NM_130435
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E
|
|
chr2_+_241375030
|
1.485
|
NM_002081
|
GPC1
|
glypican 1
|
|
chr5_+_141488314
|
1.460
|
NM_030571
|
NDFIP1
|
Nedd4 family interacting protein 1
|
|
chr1_+_160175114
|
1.456
|
NM_003768
|
PEA15
|
phosphoprotein enriched in astrocytes 15
|
|
chr3_+_36421944
|
1.456
|
NM_003149
|
STAC
|
SH3 and cysteine rich domain
|
|
chr10_-_29923900
|
1.455
|
NM_021738
|
SVIL
|
supervillin
|
|
chr3_+_37903643
|
1.439
|
NM_001008392
NM_005808
|
CTDSPL
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like
|
|
chr2_-_37899227
|
1.416
|
NM_006449
|
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3
|
|
chr17_-_7298160
|
1.408
|
NM_001201576
NM_020360
|
PLSCR3
|
phospholipid scramblase 3
|
|
chr22_+_29702950
|
1.397
|
NM_006478
NM_152236
NM_152237
|
GAS2L1
|
growth arrest-specific 2 like 1
|
|
chr10_-_3827418
|
1.372
|
NM_001160124
NM_001160125
NM_001300
|
KLF6
|
Kruppel-like factor 6
|
|
chr11_+_19734821
|
1.371
|
NM_001244963
NM_145117
NM_182964
|
NAV2
|
neuron navigator 2
|
|
chr8_-_145050896
|
1.369
|
NM_000445
|
PLEC
|
plectin
|
|
chr18_+_11751470
|
1.363
|
NM_001142339
NM_002071
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type
|
|
chr13_-_80915041
|
1.361
|
NM_005842
|
SPRY2
|
sprouty homolog 2 (Drosophila)
|
|
chr1_-_33168356
|
1.356
|
NM_001161708
NM_030786
|
SYNC
|
syncoilin, intermediate filament protein
|
|
chr3_-_133969563
|
1.320
|
NM_001005861
NM_002958
|
RYK
|
RYK receptor-like tyrosine kinase
|
|
chr19_-_10697907
|
1.319
|
NM_005498
|
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit
|
|
chr10_+_102106771
|
1.317
|
NM_005063
|
SCD
|
stearoyl-CoA desaturase (delta-9-desaturase)
|
|
chr14_+_104182045
|
1.306
|
NM_001198953
NM_024071
|
ZFYVE21
|
zinc finger, FYVE domain containing 21
|
|
chr2_-_110371669
|
1.304
|
NM_144710
NM_178584
|
SEPT10
|
septin 10
|
|
chr14_+_55034329
|
1.298
|
NM_001161576
NM_015589
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr11_-_117166247
|
1.292
|
NM_001207048
NM_001207049
|
BACE1
|
beta-site APP-cleaving enzyme 1
|
|
chr12_+_56915587
|
1.264
|
NM_002898
|
RBMS2
|
RNA binding motif, single stranded interacting protein 2
|
|
chr4_-_122618140
|
1.235
|
NM_001154
|
ANXA5
|
annexin A5
|
|
chr15_+_90931470
|
1.230
|
NM_003870
|
IQGAP1
|
IQ motif containing GTPase activating protein 1
|
|
chr10_+_123923104
|
1.229
|
NM_006997
NM_206860
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr5_+_82767492
|
1.212
|
NM_001126336
NM_001164097
NM_001164098
NM_004385
|
VCAN
|
versican
|
|
chr10_+_123748688
|
1.209
|
NM_206861
NM_206862
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr6_+_148663728
|
1.197
|
NM_015278
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr5_+_149340287
|
1.197
|
NM_000112
|
SLC26A2
|
solute carrier family 26 (sulfate transporter), member 2
|
|
chr18_+_22006605
|
1.191
|
NM_018439
|
IMPACT
|
Impact homolog (mouse)
|
|
chr17_-_41174431
|
1.181
|
NM_006373
|
VAT1
|
vesicle amine transport protein 1 homolog (T. californica)
|
|
chr11_-_117186918
|
1.172
|
NM_012104
NM_138971
NM_138972
NM_138973
|
BACE1
|
beta-site APP-cleaving enzyme 1
|
|
chr17_+_72983723
|
1.162
|
NM_014603
|
CDR2L
|
cerebellar degeneration-related protein 2-like
|
|
chr3_+_11267666
|
1.159
|
NM_001098211
|
HRH1
|
histamine receptor H1
|
|
chr11_+_19372270
|
1.156
|
NM_001111018
|
NAV2
|
neuron navigator 2
|
|
chr8_+_9911797
|
1.147
|
NM_001135670
NM_012331
|
MSRA
|
methionine sulfoxide reductase A
|
|
chr1_+_64058891
|
1.128
|
NM_002633
|
PGM1
|
phosphoglucomutase 1
|
|
chr19_-_58609606
|
1.127
|
NM_001145543
NM_001145544
NM_023926
|
ZSCAN18
|
zinc finger and SCAN domain containing 18
|
|
chr7_+_128379345
|
1.127
|
NM_001130674
NM_001199671
NM_001199672
NM_001199673
NM_001199674
NM_001219
|
CALU
|
calumenin
|
|
chr1_+_180123906
|
1.120
|
NM_001004128
NM_002826
|
QSOX1
|
quiescin Q6 sulfhydryl oxidase 1
|
|
chr2_-_230135954
|
1.113
|
NM_001100818
NM_017933
|
PID1
|
phosphotyrosine interaction domain containing 1
|
|
chr22_-_38668966
|
1.111
|
NM_001195072
NM_012264
|
TMEM184B
|
transmembrane protein 184B
|
|
chr3_+_11294384
|
1.107
|
NM_000861
|
HRH1
|
histamine receptor H1
|
|
chr10_-_91011586
|
1.103
|
NM_000235
NM_001127605
|
LIPA
|
lipase A, lysosomal acid, cholesterol esterase
|
|
chr6_+_53659738
|
1.100
|
NM_018214
|
LRRC1
|
leucine rich repeat containing 1
|
|
chr12_-_89746244
|
1.100
|
NM_001946
NM_022652
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr14_-_105635114
|
1.097
|
NM_002226
NM_145159
|
JAG2
|
jagged 2
|
|
chr22_-_38668669
|
1.085
|
NM_001195071
|
TMEM184B
|
transmembrane protein 184B
|
|
chr1_+_64088886
|
1.078
|
NM_001172818
|
PGM1
|
phosphoglucomutase 1
|
|
chr7_+_17338182
|
1.075
|
NM_001621
|
AHR
|
aryl hydrocarbon receptor
|
|
chr22_-_36784055
|
1.058
|
NM_002473
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr15_-_23086290
|
1.055
|
NM_144599
|
NIPA1
|
non imprinted in Prader-Willi/Angelman syndrome 1
|
|
chr2_+_24272583
|
1.054
|
NM_004116
NM_054033
|
FKBP1B
|
FK506 binding protein 1B, 12.6 kDa
|
|
chr2_+_131674223
|
1.043
|
NM_015320
NM_032995
|
ARHGEF4
|
Rho guanine nucleotide exchange factor (GEF) 4
|
|
chr3_-_45187897
|
1.027
|
NM_022842
NM_178181
|
CDCP1
|
CUB domain containing protein 1
|
|
chr16_-_4166185
|
1.025
|
NM_001116
|
ADCY9
|
adenylate cyclase 9
|
|
chr9_-_99329092
|
1.007
|
NM_001077181
|
CDC14B
|
CDC14 cell division cycle 14 homolog B (S. cerevisiae)
|
|
chr7_-_42276617
|
1.007
|
NM_000168
|
GLI3
|
GLI family zinc finger 3
|
|
chr18_+_11689135
|
1.002
|
NM_182978
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type
|
|
chr10_-_15413054
|
0.999
|
NM_001010924
|
FAM171A1
|
family with sequence similarity 171, member A1
|
|
chr19_-_58629762
|
0.998
|
NM_001145542
|
ZSCAN18
|
zinc finger and SCAN domain containing 18
|
|
chr4_-_40859081
|
0.997
|
NM_001166051
NM_001166052
NM_001166053
NM_001166054
|
APBB2
|
amyloid beta (A4) precursor protein-binding, family B, member 2
|
|
chr21_-_46293609
|
0.996
|
NM_004339
|
PTTG1IP
|
pituitary tumor-transforming 1 interacting protein
|
|
chr17_-_42907606
|
0.974
|
NM_005497
|
GJC1
|
gap junction protein, gamma 1, 45kDa
|
|
chr2_+_31456879
|
0.971
|
NM_014600
|
EHD3
|
EH-domain containing 3
|
|
chr8_-_57906423
|
0.962
|
NM_017813
|
IMPAD1
|
inositol monophosphatase domain containing 1
|
|
chr14_-_30396811
|
0.954
|
NM_002742
|
PRKD1
|
protein kinase D1
|
|
chr17_+_28705922
|
0.947
|
NM_001304
|
CPD
|
carboxypeptidase D
|
|
chr9_+_114423849
|
0.935
|
NM_001017998
NM_001198664
|
GNG10
|
guanine nucleotide binding protein (G protein), gamma 10
|
|
chr11_-_10562689
|
0.928
|
NM_016422
|
RNF141
|
ring finger protein 141
|
|
chr5_-_39425297
|
0.923
|
NM_001244871
NM_001343
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr4_-_120549115
|
0.919
|
NM_033437
|
PDE5A
|
phosphodiesterase 5A, cGMP-specific
|
|
chr4_-_41216455
|
0.916
|
NM_001166050
NM_004307
NM_173075
|
APBB2
|
amyloid beta (A4) precursor protein-binding, family B, member 2
|
|
chr18_-_61089695
|
0.916
|
NM_004869
|
VPS4B
|
vacuolar protein sorting 4 homolog B (S. cerevisiae)
|
|
chrX_+_66788682
|
0.915
|
NM_001011645
|
AR
|
androgen receptor
|
|
chr2_-_73340073
|
0.911
|
NM_015470
|
RAB11FIP5
|
RAB11 family interacting protein 5 (class I)
|
|
chr8_+_9953061
|
0.911
|
NM_001135671
NM_001199729
|
MSRA
|
methionine sulfoxide reductase A
|
|
chr11_-_17191287
|
0.910
|
NM_002645
|
PIK3C2A
|
phosphoinositide-3-kinase, class 2, alpha polypeptide
|
|
chr20_-_10654429
|
0.910
|
NM_000214
|
JAG1
|
jagged 1
|
|
chr3_+_50284325
|
0.903
|
NM_001166425
|
GNAI2
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2
|
|
chr19_-_48867149
|
0.899
|
NM_018273
|
TMEM143
|
transmembrane protein 143
|
|
chr1_+_220701547
|
0.899
|
NM_018650
|
MARK1
|
MAP/microtubule affinity-regulating kinase 1
|
|
chr1_+_64059479
|
0.894
|
NM_001172819
|
PGM1
|
phosphoglucomutase 1
|
|
chr3_+_105085553
|
0.893
|
NM_001243280
NM_001243281
NM_001243283
NM_001627
|
ALCAM
|
activated leukocyte cell adhesion molecule
|
|
chr11_-_115375065
|
0.879
|
NM_001098517
NM_014333
|
CADM1
|
cell adhesion molecule 1
|
|
chr17_-_42908178
|
0.875
|
NM_001080383
|
GJC1
|
gap junction protein, gamma 1, 45kDa
|
|
chr9_-_124132503
|
0.875
|
NM_004099
NM_198194
|
STOM
|
stomatin
|
|
chr8_+_95732101
|
0.875
|
NM_181787
|
DPY19L4
|
dpy-19-like 4 (C. elegans)
|
|
chr14_-_75422466
|
0.873
|
NM_001207012
NM_002632
|
PGF
|
placental growth factor
|
|
chr12_+_26348268
|
0.869
|
NM_001135823
|
SSPN
|
sarcospan (Kras oncogene-associated gene)
|
|
chr17_+_52978022
|
0.868
|
NM_005486
|
TOM1L1
|
target of myb1 (chicken)-like 1
|
|
chr17_+_68165660
|
0.867
|
NM_000891
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2
|
|
chr1_-_117664349
|
0.866
|
NM_001145635
NM_025188
|
TRIM45
|
tripartite motif containing 45
|
|
chr19_-_36523540
|
0.859
|
NM_001199570
|
CLIP3
|
CAP-GLY domain containing linker protein 3
|
|
chr2_+_70485204
|
0.854
|
NM_016297
|
PCYOX1
|
prenylcysteine oxidase 1
|
|
chr20_+_62496580
|
0.848
|
NM_001243891
NM_001243892
NM_001243894
NM_001243895
NM_003288
NM_199359
NM_199360
NM_199361
NM_199362
NM_199363
|
TPD52L2
|
tumor protein D52-like 2
|
|
chr6_+_86159301
|
0.847
|
NM_001204813
NM_002526
|
NT5E
|
5'-nucleotidase, ecto (CD73)
|
|
chr19_-_36523770
|
0.845
|
NM_015526
|
CLIP3
|
CAP-GLY domain containing linker protein 3
|
|
chr2_-_3523345
|
0.831
|
NM_018269
|
ADI1
|
acireductone dioxygenase 1
|
|
chr6_+_143999058
|
0.828
|
NM_001100164
NM_001100165
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr8_+_32579350
|
0.826
|
NM_001159996
|
NRG1
|
neuregulin 1
|
|
chr12_-_118541655
|
0.823
|
NM_019086
|
VSIG10
|
V-set and immunoglobulin domain containing 10
|
|
chr2_-_122042767
|
0.819
|
NM_014553
|
TFCP2L1
|
transcription factor CP2-like 1
|
|
chr15_+_69452799
|
0.814
|
NM_015554
|
GLCE
|
glucuronic acid epimerase
|
|
chr16_-_10674443
|
0.810
|
NM_001424
|
EMP2
|
epithelial membrane protein 2
|
|
chrX_-_134185978
|
0.807
|
NM_001078172
NM_001134321
|
FAM127B
|
family with sequence similarity 127, member B
|
|
chr9_-_99382073
|
0.807
|
NM_003671
NM_033331
|
CDC14B
|
CDC14 cell division cycle 14 homolog B (S. cerevisiae)
|
|
chr1_+_93913566
|
0.802
|
NM_001024948
NM_001164473
NM_017737
|
FNBP1L
|
formin binding protein 1-like
|