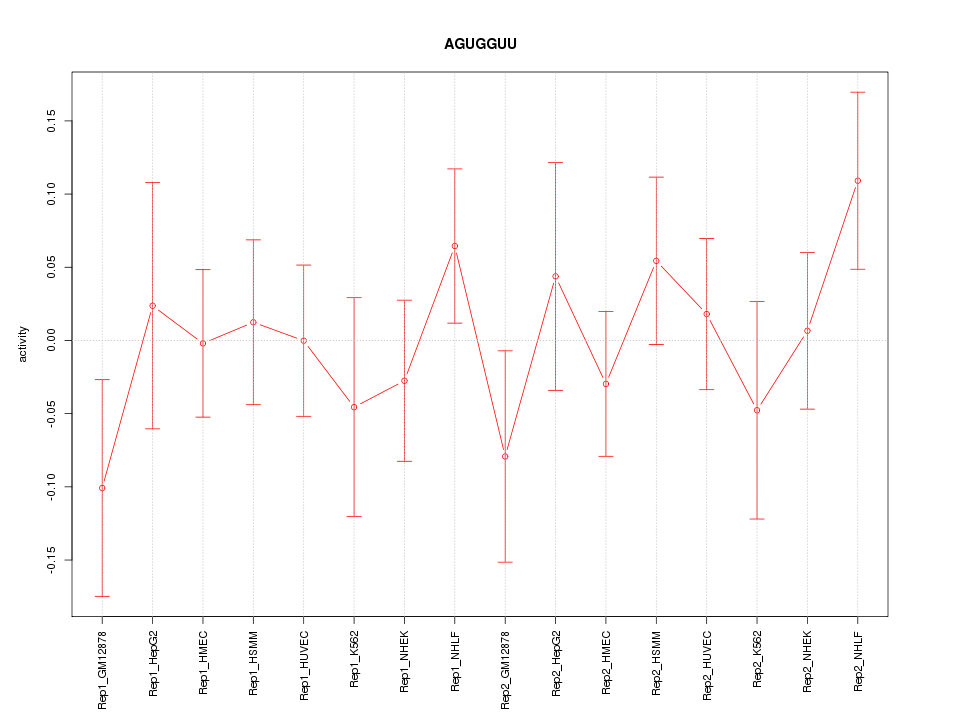
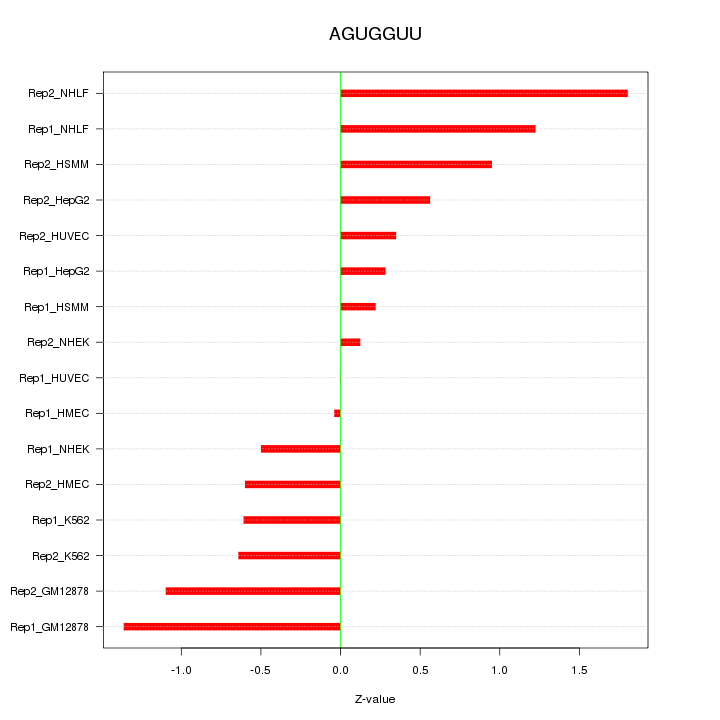
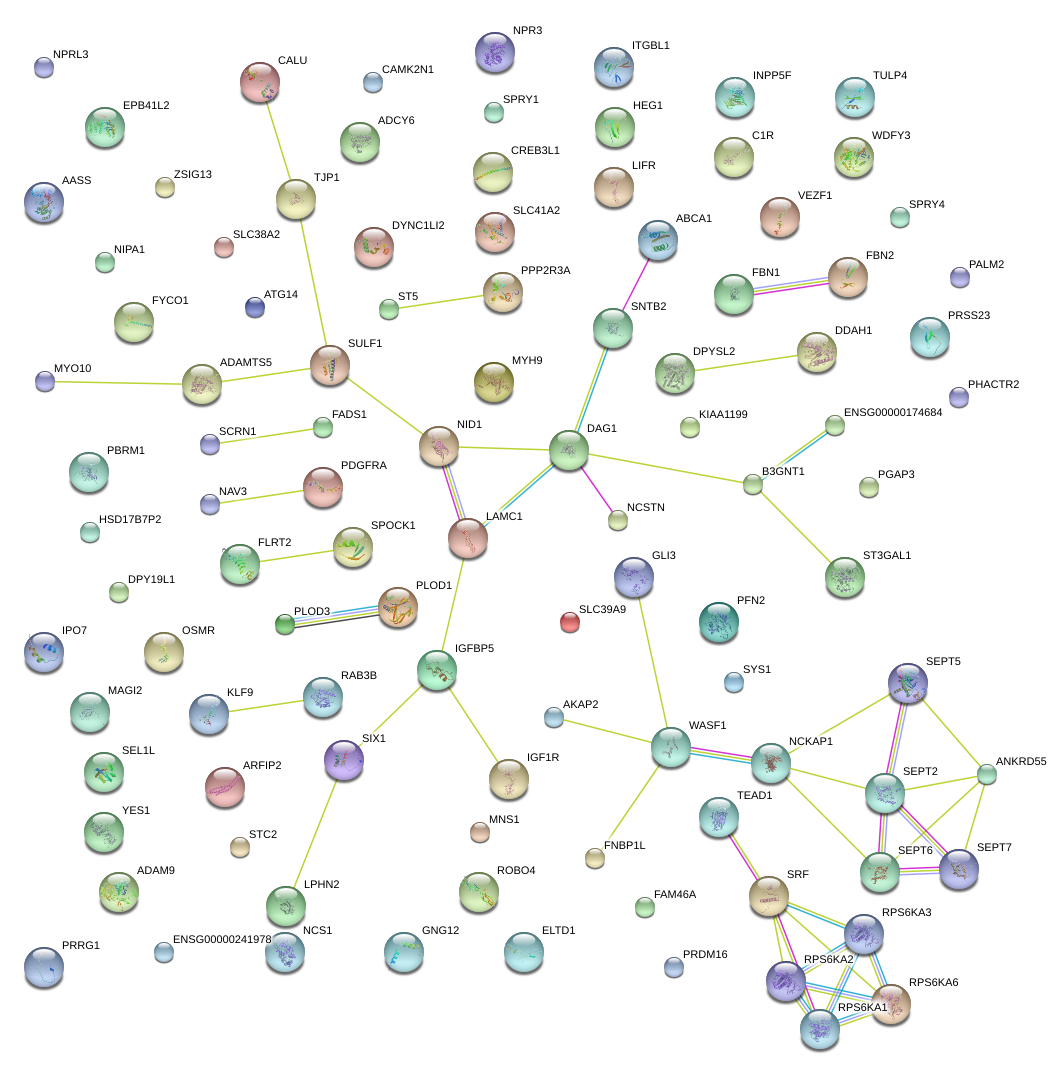
|
chr15_+_81071708
|
2.389
|
NM_018689
|
KIAA1199
|
KIAA1199
|
|
chr5_-_172756505
|
2.043
|
NM_003714
|
STC2
|
stanniocalcin 2
|
|
chr4_+_55095263
|
1.872
|
NM_006206
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr5_-_16936371
|
1.555
|
NM_012334
|
MYO10
|
myosin X
|
|
chr1_-_68299047
|
1.449
|
NM_018841
|
GNG12
|
guanine nucleotide binding protein (G protein), gamma 12
|
|
chr15_-_48937795
|
1.439
|
NM_000138
|
FBN1
|
fibrillin 1
|
|
chr2_-_217560247
|
1.431
|
NM_000599
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr11_+_86511490
|
1.331
|
NM_007173
|
PRSS23
|
protease, serine, 23
|
|
chr6_-_82462374
|
1.298
|
NM_017633
|
FAM46A
|
family with sequence similarity 46, member A
|
|
chr8_+_26435339
|
1.282
|
NM_001386
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr9_-_107690526
|
1.274
|
NM_005502
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr8_+_38854431
|
1.218
|
NM_003816
|
ADAM9
|
ADAM metallopeptidase domain 9
|
|
chr1_+_82266081
|
1.183
|
NM_012302
|
LPHN2
|
latrophilin 2
|
|
chr8_+_70405027
|
1.182
|
NM_001128205
NM_001128206
|
SULF1
|
sulfatase 1
|
|
chr1_+_182992329
|
1.177
|
NM_002293
|
LAMC1
|
laminin, gamma 1 (formerly LAMB2)
|
|
chr8_+_70378858
|
1.169
|
NM_001128204
NM_015170
|
SULF1
|
sulfatase 1
|
|
chr11_-_8932497
|
1.136
|
NM_005418
|
ST5
|
suppression of tumorigenicity 5
|
|
chr11_-_8832881
|
1.091
|
NM_139157
|
ST5
|
suppression of tumorigenicity 5
|
|
chr2_-_183903225
|
1.082
|
NM_013436
NM_205842
|
NCKAP1
|
NCK-associated protein 1
|
|
chr8_+_26371461
|
1.039
|
NM_001197293
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr16_+_69220940
|
0.974
|
NM_006750
|
SNTB2
|
syntrophin, beta 2 (dystrophin-associated protein A1, 59kDa, basic component 2)
|
|
chr5_+_32710742
|
0.968
|
NM_001204376
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C)
|
|
chr13_+_102104941
|
0.950
|
NM_004791
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains)
|
|
chr11_+_12695851
|
0.913
|
NM_021961
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor)
|
|
chr6_-_131384387
|
0.913
|
NM_001135554
NM_001135555
NM_001199388
NM_001431
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2
|
|
chr5_+_32711437
|
0.903
|
NM_000908
NM_001204375
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C)
|
|
chr11_-_8832189
|
0.890
|
NM_213618
|
ST5
|
suppression of tumorigenicity 5
|
|
chr7_-_30029904
|
0.880
|
NM_001145514
|
SCRN1
|
secernin 1
|
|
chr3_+_135684463
|
0.858
|
NM_001190447
NM_002718
|
PPP2R3A
|
protein phosphatase 2, regulatory subunit B'', alpha
|
|
chr3_+_135741576
|
0.842
|
NM_181897
|
PPP2R3A
|
protein phosphatase 2, regulatory subunit B'', alpha
|
|
chr21_-_28339405
|
0.819
|
NM_007038
|
ADAMTS5
|
ADAM metallopeptidase with thrombospondin type 1 motif, 5
|
|
chr1_-_20812727
|
0.802
|
NM_018584
|
CAMK2N1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr6_-_131291480
|
0.773
|
NM_001199389
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2
|
|
chr1_-_85930733
|
0.764
|
NM_012137
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr7_+_128379345
|
0.758
|
NM_001130674
NM_001199671
NM_001199672
NM_001199673
NM_001199674
NM_001219
|
CALU
|
calumenin
|
|
chr1_-_86043932
|
0.755
|
NM_001134445
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr6_+_143929316
|
0.743
|
NM_001100166
NM_014721
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr3_-_149688638
|
0.735
|
NM_002628
NM_053024
|
PFN2
|
profilin 2
|
|
chr6_+_143999058
|
0.729
|
NM_001100164
NM_001100165
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr6_-_110501164
|
0.717
|
NM_001024934
NM_001024935
NM_001024936
NM_003931
|
WASF1
|
WAS protein family, member 1
|
|
chr4_+_124320664
|
0.703
|
NM_005841
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila)
|
|
chr7_-_30029284
|
0.696
|
NM_001145515
NM_014766
NM_001145513
|
SCRN1
|
secernin 1
|
|
chr3_-_46037299
|
0.685
|
NM_024513
|
FYCO1
|
FYVE and coiled-coil domain containing 1
|
|
chr1_-_236228476
|
0.682
|
NM_002508
|
NID1
|
nidogen 1
|
|
chr12_+_78225068
|
0.674
|
NM_014903
|
NAV3
|
neuron navigator 3
|
|
chr3_-_124774735
|
0.666
|
NM_020733
|
HEG1
|
HEG homolog 1 (zebrafish)
|
|
chr4_+_124317884
|
0.630
|
NM_199327
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila)
|
|
chr2_+_242255270
|
0.619
|
NM_001008492
NM_004404
|
SEPT2
|
septin 2
|
|
chr9_-_73029539
|
0.593
|
NM_001206
|
KLF9
|
Kruppel-like factor 9
|
|
chr9_+_112887780
|
0.590
|
NM_001136562
|
AKAP2
|
A kinase (PRKA) anchor protein 2
|
|
chr12_-_46766563
|
0.587
|
NM_018976
|
SLC38A2
|
solute carrier family 38, member 2
|
|
chr9_+_112810877
|
0.573
|
NM_001004065
NM_001198656
|
AKAP2
|
A kinase (PRKA) anchor protein 2
|
|
chr15_-_30114536
|
0.546
|
NM_003257
NM_175610
|
TJP1
|
tight junction protein 1 (zona occludens 1)
|
|
chr5_-_127873734
|
0.544
|
NM_001999
|
FBN2
|
fibrillin 2
|
|
chr3_+_49506079
|
0.540
|
NM_001177643
|
DAG1
|
dystroglycan 1 (dystrophin-associated glycoprotein 1)
|
|
chr9_+_112542496
|
0.533
|
NM_053016
NM_007203
NM_147150
|
PALM2
PALM2-AKAP2
|
paralemmin 2
PALM2-AKAP2 readthrough
|
|
chr1_+_11994710
|
0.513
|
NM_000302
|
PLOD1
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1
|
|
chr1_-_52456292
|
0.501
|
NM_002867
|
RAB3B
|
RAB3B, member RAS oncogene family
|
|
chr1_-_79472360
|
0.495
|
NM_022159
|
ELTD1
|
EGF, latrophilin and seven transmembrane domain containing 1
|
|
chr16_-_66785511
|
0.488
|
NM_006141
|
DYNC1LI2
|
dynein, cytoplasmic 1, light intermediate chain 2
|
|
chrX_+_37208527
|
0.483
|
NM_000950
NM_001142395
NM_001173486
NM_001173489
NM_001173490
|
PRRG1
|
proline rich Gla (G-carboxyglutamic acid) 1
|
|
chr7_-_121784236
|
0.482
|
NM_005763
|
AASS
|
aminoadipate-semialdehyde synthase
|
|
chr11_-_66115016
|
0.478
|
NM_006876
|
B3GNT1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1
|
|
chr17_-_37844266
|
0.477
|
NM_033419
|
PGAP3
|
post-GPI attachment to proteins 3
|
|
chr5_-_38595506
|
0.470
|
NM_002310
|
LIFR
|
leukemia inhibitory factor receptor alpha
|
|
chr15_-_23086290
|
0.457
|
NM_144599
|
NIPA1
|
non imprinted in Prader-Willi/Angelman syndrome 1
|
|
chr15_+_99192696
|
0.449
|
NM_000875
|
IGF1R
|
insulin-like growth factor 1 receptor
|
|
chr5_-_141704575
|
0.439
|
NM_001127496
NM_030964
|
SPRY4
|
sprouty homolog 4 (Drosophila)
|
|
chr4_-_85887543
|
0.435
|
NM_014991
|
WDFY3
|
WD repeat and FYVE domain containing 3
|
|
chr5_+_38845962
|
0.429
|
NM_003999
|
OSMR
|
oncostatin M receptor
|
|
chr2_+_242254722
|
0.426
|
NM_001008491
NM_006155
|
SEPT2
|
septin 2
|
|
chr14_+_85996454
|
0.417
|
NM_013231
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr12_-_105322056
|
0.415
|
NM_032148
|
SLC41A2
|
solute carrier family 41, member 2
|
|
chr18_-_812268
|
0.409
|
NM_005433
|
YES1
|
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1
|
|
chr7_-_42276617
|
0.408
|
NM_000168
|
GLI3
|
GLI family zinc finger 3
|
|
chr14_-_61115906
|
0.408
|
NM_005982
|
SIX1
|
SIX homeobox 1
|
|
chr12_-_49177876
|
0.393
|
NM_015270
|
ADCY6
|
adenylate cyclase 6
|
|
chr3_+_49507470
|
0.381
|
NM_001165928
NM_001177634
NM_001177635
NM_001177636
NM_001177637
NM_001177638
NM_001177639
NM_001177640
NM_001177641
NM_001177642
NM_001177644
NM_004393
|
DAG1
|
dystroglycan 1 (dystrophin-associated glycoprotein 1)
|
|
chr9_+_132962871
|
0.380
|
NM_001128826
|
NCS1
|
neuronal calcium sensor 1
|
|
chr14_-_82000198
|
0.366
|
NM_001244984
NM_005065
|
SEL1L
|
sel-1 suppressor of lin-12-like (C. elegans)
|
|
chr8_-_134584012
|
0.365
|
NM_003033
NM_173344
|
ST3GAL1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1
|
|
chr14_-_55878537
|
0.356
|
NM_014924
|
ATG14
|
ATG14 autophagy related 14 homolog (S. cerevisiae)
|
|
chr12_-_7245006
|
0.354
|
NM_001733
|
C1R
|
complement component 1, r subcomponent
|
|
chr7_-_79082870
|
0.353
|
NM_012301
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2
|
|
chr11_+_46299161
|
0.353
|
NM_052854
|
CREB3L1
|
cAMP responsive element binding protein 3-like 1
|
|
chr11_+_9406123
|
0.352
|
NM_006391
|
IPO7
|
importin 7
|
|
chr15_-_56757239
|
0.350
|
NM_018365
|
MNS1
|
meiosis-specific nuclear structural 1
|
|
chr6_+_158733691
|
0.344
|
NM_001007466
NM_020245
|
TULP4
|
tubby like protein 4
|
|
chr5_-_136834887
|
0.341
|
NM_004598
|
SPOCK1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1
|
|
chr11_-_61584451
|
0.338
|
NM_013402
|
FADS1
|
fatty acid desaturase 1
|
|
chr11_-_6502563
|
0.329
|
NM_001242854
NM_001242855
NM_001242856
NM_012402
|
ARFIP2
|
ADP-ribosylation factor interacting protein 2
|
|
chr20_+_43991739
|
0.325
|
NM_001099791
NM_033542
|
SYS1-DBNDD2
SYS1
|
SYS1-DBNDD2 readthrough
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae)
|
|
chr7_-_100860991
|
0.322
|
NM_001084
|
PLOD3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3
|
|
chr1_+_160313061
|
0.312
|
NM_015331
|
NCSTN
|
nicastrin
|
|
chr17_-_56065596
|
0.308
|
NM_007146
|
VEZF1
|
vascular endothelial zinc finger 1
|
|
chr14_+_69865380
|
0.302
|
NM_001252148
NM_001252150
NM_018375
|
SLC39A9
|
solute carrier family 39 (zinc transporter), member 9
|
|
chr3_-_52719514
|
0.299
|
NM_018313
|
PBRM1
|
polybromo 1
|
|
chr7_-_35077652
|
0.299
|
NM_015283
|
DPY19L1
|
dpy-19-like 1 (C. elegans)
|
|
chr1_+_93913566
|
0.297
|
NM_001024948
NM_001164473
NM_017737
|
FNBP1L
|
formin binding protein 1-like
|
|
chr22_-_36784055
|
0.295
|
NM_002473
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chrX_-_20284708
|
0.294
|
NM_004586
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3
|
|
chr9_+_132815984
|
0.292
|
NM_001136557
NM_001136558
NM_020960
|
GPR107
|
G protein-coupled receptor 107
|
|
chr17_+_61086897
|
0.292
|
NM_025185
|
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2
|
|
chr1_-_168105620
|
0.277
|
NM_153832
|
GPR161
|
G protein-coupled receptor 161
|
|
chr2_-_240322620
|
0.274
|
NM_006037
|
HDAC4
|
histone deacetylase 4
|
|
chr4_-_111563278
|
0.270
|
NM_001204397
NM_001204398
NM_001204399
NM_153426
NM_153427
|
PITX2
|
paired-like homeodomain 2
|
|
chr6_+_57037058
|
0.267
|
NM_004282
|
BAG2
|
BCL2-associated athanogene 2
|
|
chr14_+_56584854
|
0.251
|
NM_021255
|
PELI2
|
pellino homolog 2 (Drosophila)
|
|
chr5_-_38556682
|
0.242
|
NM_001127671
|
LIFR
|
leukemia inhibitory factor receptor alpha
|
|
chr8_-_67579446
|
0.239
|
NM_025054
|
VCPIP1
|
valosin containing protein (p97)/p47 complex interacting protein 1
|
|
chr19_+_11200037
|
0.235
|
NM_000527
NM_001195798
NM_001195799
NM_001195800
NM_001195802
NM_001195803
|
LDLR
|
low density lipoprotein receptor
|
|
chr10_+_31608097
|
0.235
|
NM_001174093
NM_001174095
NM_001174096
NM_030751
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr6_-_47277646
|
0.235
|
NM_014452
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21
|
|
chr3_-_50396930
|
0.234
|
NM_007024
|
TMEM115
|
transmembrane protein 115
|
|
chr15_-_59041933
|
0.231
|
NM_001110
|
ADAM10
|
ADAM metallopeptidase domain 10
|
|
chr4_-_111544206
|
0.229
|
NM_000325
|
PITX2
|
paired-like homeodomain 2
|
|
chr10_-_104262404
|
0.226
|
NM_005736
|
ACTR1A
|
ARP1 actin-related protein 1 homolog A, centractin alpha (yeast)
|
|
chr11_-_47270339
|
0.224
|
NM_001131064
NM_001610
|
ACP2
|
acid phosphatase 2, lysosomal
|
|
chr14_+_65171149
|
0.222
|
NM_015549
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3
|
|
chr16_-_48419148
|
0.222
|
NM_003031
|
SIAH1
|
seven in absentia homolog 1 (Drosophila)
|
|
chr13_-_110959441
|
0.222
|
NM_001845
|
COL4A1
|
collagen, type IV, alpha 1
|
|
chr21_-_15755439
|
0.219
|
NM_006948
|
HSPA13
|
heat shock protein 70kDa family, member 13
|
|
chr5_-_77944341
|
0.217
|
NM_005779
|
LHFPL2
|
lipoma HMGIC fusion partner-like 2
|
|
chr7_+_94539209
|
0.212
|
NM_001166161
NM_001166162
NM_001166163
|
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A
|
|
chr6_-_108395939
|
0.212
|
NM_014028
|
OSTM1
|
osteopetrosis associated transmembrane protein 1
|
|
chrY_+_15016018
|
0.203
|
NM_001122665
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked
|
|
chr2_+_5832778
|
0.203
|
NM_003108
|
SOX11
|
SRY (sex determining region Y)-box 11
|
|
chr20_-_10654429
|
0.200
|
NM_000214
|
JAG1
|
jagged 1
|
|
chrX_+_107069081
|
0.199
|
NM_012216
NM_052817
|
MID2
|
midline 2
|
|
chr1_+_15272414
|
0.198
|
NM_001017999
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr7_-_32931409
|
0.197
|
NM_015483
|
KBTBD2
|
kelch repeat and BTB (POZ) domain containing 2
|
|
chr4_+_123747815
|
0.196
|
NM_002006
|
FGF2
|
fibroblast growth factor 2 (basic)
|
|
chr10_+_104503724
|
0.192
|
NM_001083913
|
C10orf26
|
chromosome 10 open reading frame 26
|
|
chr3_+_32148000
|
0.191
|
NM_015141
|
GPD1L
|
glycerol-3-phosphate dehydrogenase 1-like
|
|
chr16_+_83841572
|
0.180
|
NM_001537
|
HSBP1
|
heat shock factor binding protein 1
|
|
chr10_+_31610063
|
0.179
|
NM_001128128
NM_001174094
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr12_-_49182718
|
0.176
|
NM_020983
|
ADCY6
|
adenylate cyclase 6
|
|
chr1_-_146644045
|
0.174
|
NM_005399
|
PRKAB2
|
protein kinase, AMP-activated, beta 2 non-catalytic subunit
|
|
chr10_+_102672325
|
0.172
|
NM_001136123
NM_001243770
NM_018121
|
FAM178A
|
family with sequence similarity 178, member A
|
|
chr9_-_79009265
|
0.167
|
NM_018339
|
RFK
|
riboflavin kinase
|
|
chrY_+_15016698
|
0.166
|
NM_004660
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked
|
|
chr3_+_33318911
|
0.166
|
NM_012157
NM_001171713
|
FBXL2
|
F-box and leucine-rich repeat protein 2
|
|
chr22_+_25202135
|
0.164
|
NM_001039948
NM_001098497
NM_001098498
NM_133454
|
SGSM1
|
small G protein signaling modulator 1
|
|
chr16_-_48399783
|
0.164
|
NM_001006610
|
SIAH1
|
seven in absentia homolog 1 (Drosophila)
|
|
chr1_+_24742244
|
0.163
|
NM_020448
|
NIPAL3
|
NIPA-like domain containing 3
|
|
chr3_+_15469052
|
0.161
|
NM_033083
|
EAF1
|
ELL associated factor 1
|
|
chr16_-_2264776
|
0.154
|
NM_001042371
|
PGP
|
phosphoglycolate phosphatase
|
|
chr14_+_57046490
|
0.153
|
NM_017799
|
C14orf101
|
chromosome 14 open reading frame 101
|
|
chr14_+_23775970
|
0.151
|
NM_001199864
NM_004050
NM_001199839
|
BCL2L2-PABPN1
BCL2L2
|
BCL2L2-PABPN1 readthrough
BCL2-like 2
|
|
chr5_-_9546157
|
0.150
|
NM_003966
|
SEMA5A
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A
|
|
chr2_-_178129858
|
0.150
|
NM_006164
|
NFE2L2
|
nuclear factor (erythroid-derived 2)-like 2
|
|
chr3_+_150126705
|
0.150
|
NM_014779
|
TSC22D2
|
TSC22 domain family, member 2
|
|
chr19_-_47735858
|
0.145
|
NM_001127240
NM_001127241
NM_001127242
|
BBC3
|
BCL2 binding component 3
|
|
chr1_+_78470593
|
0.141
|
NM_007034
|
DNAJB4
|
DnaJ (Hsp40) homolog, subfamily B, member 4
|
|
chr9_+_132934685
|
0.141
|
NM_014286
|
NCS1
|
neuronal calcium sensor 1
|
|
chr11_-_46939908
|
0.137
|
NM_002334
|
LRP4
|
low density lipoprotein receptor-related protein 4
|
|
chr19_-_49496566
|
0.136
|
NM_001161587
NM_002103
|
GYS1
|
glycogen synthase 1 (muscle)
|
|
chr3_+_42947401
|
0.136
|
NM_207404
|
ZNF662
|
zinc finger protein 662
|
|
chrX_-_119603155
|
0.134
|
NM_001122606
NM_002294
NM_013995
|
LAMP2
|
lysosomal-associated membrane protein 2
|
|
chr12_-_27167332
|
0.131
|
NM_016551
|
TM7SF3
|
transmembrane 7 superfamily member 3
|
|
chr5_+_155753755
|
0.131
|
NM_000337
NM_001128209
NM_172244
|
SGCD
|
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein)
|
|
chr10_-_116286661
|
0.131
|
NM_006720
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr9_-_99801365
|
0.129
|
NM_001333
|
CTSL2
|
cathepsin L2
|
|
chr1_+_15256295
|
0.128
|
NM_001018001
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr2_-_178128527
|
0.127
|
NM_001145412
NM_001145413
|
NFE2L2
|
nuclear factor (erythroid-derived 2)-like 2
|
|
chr4_+_113739203
|
0.126
|
NM_001127493
|
ANK2
|
ankyrin 2, neuronal
|
|
chr19_+_48972464
|
0.124
|
NM_004228
NM_017457
|
CYTH2
|
cytohesin 2
|
|
chr9_+_120466453
|
0.124
|
NM_138554
|
TLR4
|
toll-like receptor 4
|
|
chr7_-_135662194
|
0.123
|
NM_145808
NM_001128619
|
MTPN
LUZP6
|
myotrophin
leucine zipper protein 6
|
|
chr7_+_39663140
|
0.119
|
NM_005402
|
RALA
|
v-ral simian leukemia viral oncogene homolog A (ras related)
|
|
chr1_+_204797774
|
0.118
|
NM_001005388
NM_001005389
NM_001160332
NM_001160333
NM_015090
|
NFASC
|
neurofascin
|
|
chr14_-_88460008
|
0.117
|
NM_001201402
|
GALC
|
galactosylceramidase
|
|
chr3_+_42947649
|
0.117
|
NM_001134656
|
ZNF662
|
zinc finger protein 662
|
|
chr10_-_7661613
|
0.115
|
NM_032817
|
ITIH5
|
inter-alpha-trypsin inhibitor heavy chain family, member 5
|
|
chr10_-_3827418
|
0.113
|
NM_001160124
NM_001160125
NM_001300
|
KLF6
|
Kruppel-like factor 6
|
|
chrX_+_70586113
|
0.111
|
NM_004606
NM_138923
|
TAF1
|
TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 250kDa
|
|
chr9_+_108320382
|
0.111
|
NM_001079802
NM_001198963
NM_006731
|
FKTN
|
fukutin
|
|
chr20_+_327329
|
0.109
|
NM_024958
|
NRSN2
|
neurensin 2
|
|
chr15_+_91427664
|
0.109
|
NM_001143785
NM_002005
|
FES
|
feline sarcoma oncogene
|
|
chr17_-_38256905
|
0.107
|
NM_021724
|
NR1D1
|
nuclear receptor subfamily 1, group D, member 1
|
|
chr19_+_54926604
|
0.106
|
NM_001005367
NM_001201461
NM_020659
|
TTYH1
|
tweety homolog 1 (Drosophila)
|
|
chr16_+_30710461
|
0.105
|
NM_006662
|
SRCAP
|
Snf2-related CREBBP activator protein
|
|
chrX_+_47077426
|
0.105
|
NM_033018
|
CDK16
|
cyclin-dependent kinase 16
|
|
chr17_-_58603492
|
0.103
|
NM_006380
|
APPBP2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2
|
|
chrX_-_83442906
|
0.103
|
NM_014496
|
RPS6KA6
|
ribosomal protein S6 kinase, 90kDa, polypeptide 6
|
|
chr3_+_134204863
|
0.098
|
NM_001042400
|
CEP63
|
centrosomal protein 63kDa
|
|
chr1_+_198125890
|
0.096
|
NM_133494
|
NEK7
|
NIMA (never in mitosis gene a)-related kinase 7
|
|
chr1_-_222885653
|
0.095
|
NM_022831
|
AIDA
|
axin interactor, dorsalization associated
|
|
chr6_+_52935770
|
0.093
|
NM_012347
|
FBXO9
|
F-box protein 9
|
|
chr17_+_36507708
|
0.091
|
NM_014598
|
SOCS7
|
suppressor of cytokine signaling 7
|
|
chrX_+_47082416
|
0.088
|
NM_001170460
|
CDK16
|
cyclin-dependent kinase 16
|
|
chr1_+_184356128
|
0.087
|
NM_030806
|
C1orf21
|
chromosome 1 open reading frame 21
|
|
chr1_+_15250578
|
0.086
|
NM_001018000
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr14_-_90085325
|
0.085
|
NM_001085471
|
FOXN3
|
forkhead box N3
|
|
chr7_+_86974928
|
0.084
|
NM_001143935
NM_001243745
NM_021151
|
CROT
|
carnitine O-octanoyltransferase
|
|
chr9_-_99801924
|
0.081
|
NM_001201575
|
CTSL2
|
cathepsin L2
|
|
chr17_+_18684579
|
0.081
|
NM_016078
|
FAM18B1
|
family with sequence similarity 18, member B1
|
|
chr22_-_38714088
|
0.081
|
NM_001894
|
CSNK1E
|
casein kinase 1, epsilon
|
|
chr12_-_80307690
|
0.080
|
NM_001143886
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A
|
|
chr3_+_134204557
|
0.079
|
NM_001042383
NM_001042384
|
CEP63
|
centrosomal protein 63kDa
|