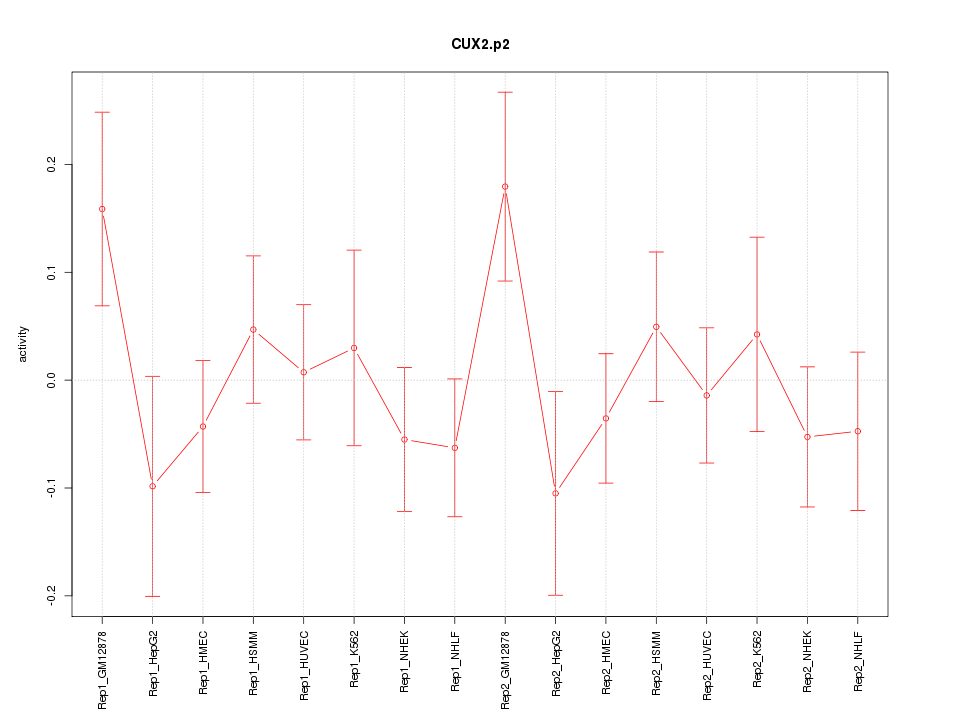
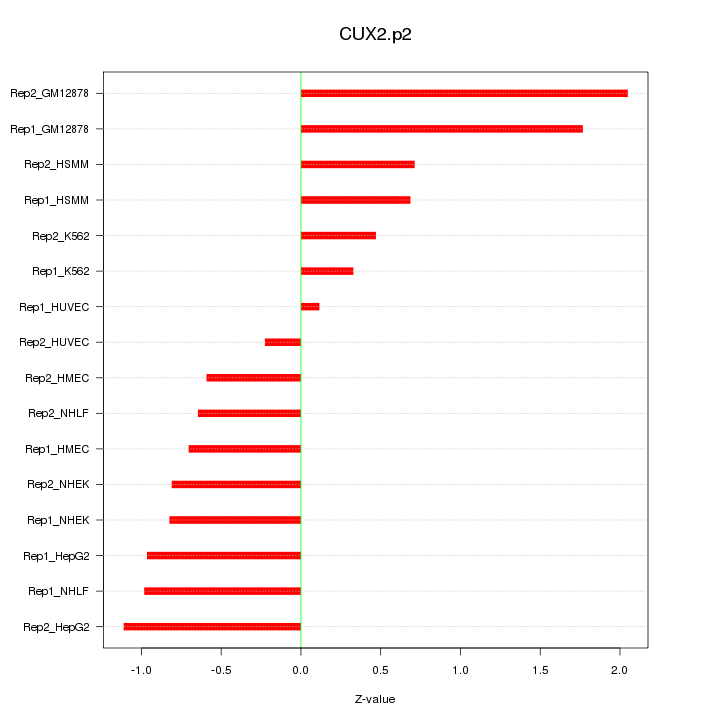
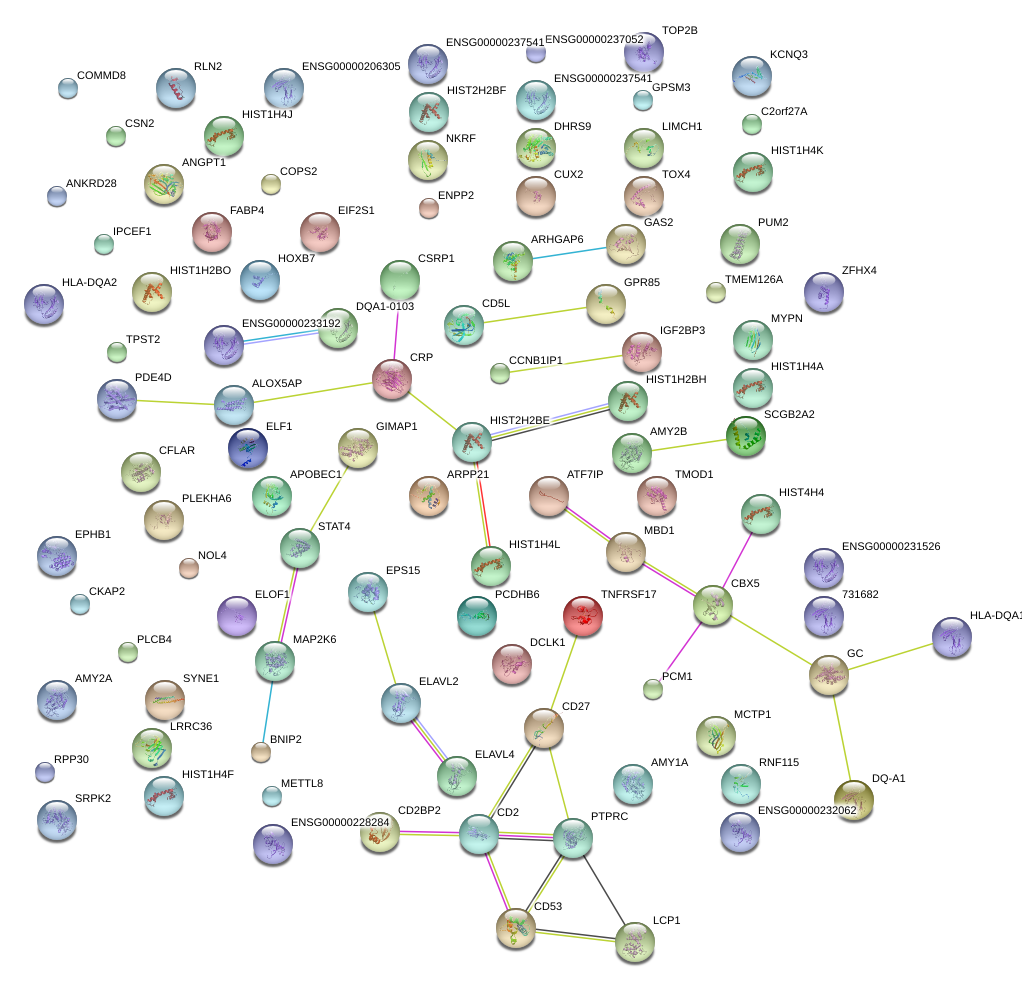
|
chr2_-_89165652
|
5.062
|
|
|
|
|
chr13_-_46756295
|
4.705
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr1_+_198608136
|
3.819
|
NM_002838
NM_080921
NM_080923
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr13_-_46756291
|
3.457
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr1_+_198608216
|
3.305
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr1_+_198608244
|
2.745
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr16_+_12058963
|
1.639
|
NM_001192
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17
|
|
chr13_-_46756326
|
1.572
|
NM_002298
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr1_-_51887740
|
1.201
|
NM_001159969
|
EPS15
|
epidermal growth factor receptor pathway substrate 15
|
|
chr6_+_32709162
|
1.190
|
NM_020056
|
HLA-DQA1
HLA-DQA2
|
major histocompatibility complex, class II, DQ alpha 1
major histocompatibility complex, class II, DQ alpha 2
|
|
chr12_-_54653317
|
1.082
|
NM_001127322
|
CBX5
|
chromobox homolog 5
|
|
chr12_+_6554032
|
1.014
|
NM_001242
|
CD27
|
CD27 molecule
|
|
chr9_-_5304579
|
0.981
|
NM_005059
NM_134441
|
RLN2
|
relaxin 2
|
|
chr2_+_202004997
|
0.951
|
NM_001202515
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator
|
|
chr12_-_54653369
|
0.906
|
NM_001127321
|
CBX5
|
chromobox homolog 5
|
|
chr17_-_46687925
|
0.835
|
|
HOXB7
|
homeobox B7
|
|
chr6_-_154677868
|
0.810
|
NM_001130700
NM_015553
|
IPCEF1
|
interaction protein for cytohesin exchange factors 1
|
|
chr1_+_111413820
|
0.770
|
NM_001040033
|
CD53
|
CD53 molecule
|
|
chr10_+_92631241
|
0.763
|
|
RPP30
|
ribonuclease P/MRP 30kDa subunit
|
|
chr13_+_53035227
|
0.751
|
|
CKAP2
|
cytoskeleton associated protein 2
|
|
chr2_+_132480063
|
0.738
|
NM_013310
|
C2orf27A
|
chromosome 2 open reading frame 27A
|
|
chr22_-_26961333
|
0.737
|
NM_001008566
|
TPST2
|
tyrosylprotein sulfotransferase 2
|
|
chr2_+_169926084
|
0.674
|
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9
|
|
chr4_-_72649734
|
0.638
|
NM_000583
|
GC
|
group-specific component (vitamin D binding protein)
|
|
chr13_+_31309644
|
0.604
|
NM_001629
|
ALOX5AP
|
arachidonate 5-lipoxygenase-activating protein
|
|
chr17_+_67498391
|
0.599
|
|
MAP2K6
|
mitogen-activated protein kinase kinase 6
|
|
chr17_-_46688362
|
0.546
|
NM_004502
|
HOXB7
|
homeobox B7
|
|
chr9_+_100263461
|
0.538
|
NM_001166116
|
TMOD1
|
tropomodulin 1
|
|
chr11_+_22689653
|
0.526
|
NM_177553
|
GAS2
|
growth arrest-specific 2
|
|
chr13_-_36429798
|
0.510
|
NM_001195415
NM_001195416
NM_001195430
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr5_-_94224601
|
0.509
|
|
MCTP1
|
multiple C2 domains, transmembrane 1
|
|
chr12_+_14538232
|
0.465
|
|
ATF7IP
|
activating transcription factor 7 interacting protein
|
|
chr8_-_120651019
|
0.451
|
NM_001040092
NM_001130863
NM_006209
|
ENPP2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2
|
|
chr3_+_35721115
|
0.449
|
NM_001025068
NM_001025069
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr1_-_159684274
|
0.448
|
NM_000567
|
CRP
|
C-reactive protein, pentraxin-related
|
|
chr1_+_50571963
|
0.438
|
NM_001144775
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr1_+_104159953
|
0.436
|
NM_000699
|
AMY2A
|
amylase, alpha 2A (pancreatic)
|
|
chr2_-_192016293
|
0.383
|
NM_001243835
|
STAT4
|
signal transducer and activator of transcription 4
|
|
chr4_-_70826712
|
0.364
|
NM_001891
|
CSN2
|
casein beta
|
|
chr8_+_77593506
|
0.357
|
NM_024721
|
ZFHX4
|
zinc finger homeobox 4
|
|
chr3_-_15839674
|
0.352
|
NM_001195098
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr8_+_77593522
|
0.349
|
|
ZFHX4
|
zinc finger homeobox 4
|
|
chr3_+_134514044
|
0.348
|
NM_004441
|
EPHB1
|
EPH receptor B1
|
|
chr6_-_152958533
|
0.317
|
NM_182961
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1
|
|
chr7_-_104909442
|
0.315
|
NM_182691
|
SRPK2
|
SRSF protein kinase 2
|
|
chr1_+_145663156
|
0.290
|
|
RNF115
|
ring finger protein 115
|
|
chrX_-_11284094
|
0.289
|
NM_013423
|
ARHGAP6
|
Rho GTPase activating protein 6
|
|
chr2_-_172291227
|
0.286
|
NM_024770
|
METTL8
|
methyltransferase like 8
|
|
chr3_-_15798057
|
0.279
|
NM_001195099
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr1_-_157811491
|
0.277
|
NM_005894
|
CD5L
|
CD5 molecule-like
|
|
chr8_-_108510094
|
0.271
|
NM_001146
NM_001199859
|
ANGPT1
|
angiopoietin 1
|
|
chr12_-_7818467
|
0.236
|
NM_001644
|
APOBEC1
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1
|
|
chr4_-_47465719
|
0.232
|
|
COMMD8
|
COMM domain containing 8
|
|
chr7_-_23509972
|
0.218
|
NM_006547
|
IGF2BP3
|
insulin-like growth factor 2 mRNA binding protein 3
|
|
chr14_+_67831484
|
0.218
|
|
EIF2S1
|
eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa
|
|
chr17_-_46688279
|
0.199
|
|
HOXB7
|
homeobox B7
|
|
chr1_-_149399159
|
0.190
|
NM_001024599
|
HIST2H2BF
|
histone cluster 2, H2bf
|
|
chr12_+_21168629
|
0.181
|
NM_001009562
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional)
|
|
chr5_+_140529619
|
0.167
|
NM_018939
|
PCDHB6
|
protocadherin beta 6
|
|
chr3_-_25683825
|
0.166
|
|
TOP2B
|
topoisomerase (DNA) II beta 180kDa
|
|
chr12_+_74931550
|
0.158
|
NM_001136262
|
ATXN7L3B
|
ataxin 7-like 3B
|
|
chr4_+_41700736
|
0.154
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chrX_-_118726852
|
0.151
|
NM_001173488
|
NKRF
|
NFKB repressing factor
|
|
chr8_-_82395401
|
0.150
|
NM_001442
|
FABP4
|
fatty acid binding protein 4, adipocyte
|
|
chr8_-_133459508
|
0.144
|
NM_001204824
|
KCNQ3
|
potassium voltage-gated channel, KQT-like subfamily, member 3
|
|
chr15_-_59981433
|
0.134
|
|
BNIP2
|
BCL2/adenovirus E1B 19kDa interacting protein 2
|
|
chr10_+_69869249
|
0.133
|
NM_032578
|
MYPN
|
myopalladin
|
|
chr8_+_17822093
|
0.132
|
|
PCM1
|
pericentriolar material 1
|
|
chr13_-_41556360
|
0.127
|
NM_001145353
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr7_-_112727832
|
0.124
|
NM_001146265
NM_018970
|
GPR85
|
G protein-coupled receptor 85
|
|
chr1_-_204329043
|
0.119
|
NM_014935
|
PLEKHA6
|
pleckstrin homology domain containing, family A member 6
|
|
chr18_-_31628557
|
0.119
|
NM_001198549
|
NOL4
|
nucleolar protein 4
|
|
chr1_+_117297056
|
0.118
|
NM_001767
|
CD2
|
CD2 molecule
|
|
chr5_-_58882274
|
0.112
|
NM_006203
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr12_+_111471827
|
0.110
|
NM_015267
|
CUX2
|
cut-like homeobox 2
|
|
chr6_-_27799304
|
0.107
|
NM_003541
|
HIST1H4A
HIST1H4F
HIST1H4K
HIST1H4J
|
histone cluster 1, H4a
histone cluster 1, H4f
histone cluster 1, H4k
histone cluster 1, H4j
|
|
chr13_-_41556188
|
0.091
|
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr16_+_67381252
|
0.083
|
NM_001161575
|
LRRC36
|
leucine rich repeat containing 36
|
|
chr11_+_62037629
|
0.082
|
NM_002411
|
SCGB2A2
|
secretoglobin, family 2A, member 2
|
|
chr6_-_32160637
|
0.081
|
|
GPSM3
|
G-protein signaling modulator 3
|
|
chr2_-_20512016
|
0.080
|
|
PUM2
|
pumilio homolog 2 (Drosophila)
|
|
chr20_+_9198036
|
0.079
|
NM_182797
|
PLCB4
|
phospholipase C, beta 4
|
|
chr7_+_150413597
|
0.075
|
NM_001199577
NM_130759
|
GIMAP1-GIMAP5
GIMAP1
|
GIMAP1-GIMAP5 readthrough
GTPase, IMAP family member 1
|
|
chr14_-_20797532
|
0.073
|
NM_182852
NM_182851
|
CCNB1IP1
|
cyclin B1 interacting protein 1, E3 ubiquitin protein ligase
|
|
chr8_+_17829848
|
0.073
|
|
PCM1
|
pericentriolar material 1
|
|
chr17_+_7461591
|
0.069
|
NM_001198622
NM_001198623
NM_001198624
NM_003808
NM_172087
NM_172088
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13
|
|
chr1_-_186430216
|
0.067
|
NM_002597
|
PDC
|
phosducin
|
|
chr6_+_129204285
|
0.066
|
NM_000426
NM_001079823
|
LAMA2
|
laminin, alpha 2
|
|
chr6_-_24719286
|
0.062
|
NM_030939
|
C6orf62
|
chromosome 6 open reading frame 62
|
|
chr20_-_17539480
|
0.058
|
NM_001161705
|
BFSP1
|
beaded filament structural protein 1, filensin
|
|
chr12_-_22089627
|
0.057
|
NM_005691
NM_020297
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9
|
|
chr4_-_47983430
|
0.055
|
NM_001142564
|
CNGA1
|
cyclic nucleotide gated channel alpha 1
|
|
chr17_-_67311773
|
0.054
|
NM_018672
|
ABCA5
|
ATP-binding cassette, sub-family A (ABC1), member 5
|
|
chr7_-_112727799
|
0.051
|
|
GPR85
|
G protein-coupled receptor 85
|
|
chr7_+_99425635
|
0.049
|
NM_022820
NM_057095
NM_057096
|
CYP3A43
|
cytochrome P450, family 3, subfamily A, polypeptide 43
|
|
chr1_+_12976449
|
0.047
|
NM_001012276
|
PRAMEF8
PRAMEF7
|
PRAME family member 8
PRAME family member 7
|
|
chr7_+_93535819
|
0.047
|
NM_021955
|
GNGT1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1
|
|
chr15_+_80364902
|
0.045
|
NM_001242915
NM_001242916
|
ZFAND6
|
zinc finger, AN1-type domain 6
|
|
chr5_+_140247767
|
0.044
|
NM_018902
NM_031861
|
PCDHA11
|
protocadherin alpha 11
|
|
chr11_-_30605748
|
0.042
|
|
MPPED2
|
metallophosphoesterase domain containing 2
|
|
chr1_-_104238785
|
0.037
|
NM_004038
NM_001008218
NM_001008219
|
AMY1A
AMY1B
AMY1C
|
amylase, alpha 1A (salivary)
amylase, alpha 1B (salivary)
amylase, alpha 1C (salivary)
|
|
chrX_-_135338640
|
0.033
|
NM_001173516
|
MAP7D3
|
MAP7 domain containing 3
|
|
chr9_-_5339745
|
0.029
|
NM_006911
|
RLN1
|
relaxin 1
|
|
chr1_+_145883867
|
0.025
|
NM_001097616
|
GPR89C
|
G protein-coupled receptor 89C
|
|
chr2_-_163008913
|
0.023
|
NM_002054
|
GCG
|
glucagon
|
|
chr12_+_52056547
|
0.019
|
NM_001177984
|
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit
|
|
chr5_-_140895407
|
0.014
|
|
DIAPH1
|
diaphanous homolog 1 (Drosophila)
|
|
chr10_+_95517565
|
0.012
|
NM_005097
|
LGI1
|
leucine-rich, glioma inactivated 1
|
|
chr2_-_77749335
|
0.011
|
NM_001134745
NM_024993
|
LRRTM4
|
leucine rich repeat transmembrane neuronal 4
|
|
chr12_+_26126687
|
0.011
|
NM_001164747
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8
|
|
chr10_-_116392099
|
0.011
|
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr6_+_29426130
|
0.010
|
NM_030883
|
OR2H1
|
olfactory receptor, family 2, subfamily H, member 1
|
|
chr17_+_56315786
|
0.008
|
NM_001160102
NM_006151
|
LPO
|
lactoperoxidase
|
|
chr21_-_31538970
|
0.007
|
NM_012131
|
CLDN17
|
claudin 17
|
|
chr2_-_152830448
|
0.007
|
NM_001005747
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr4_+_166131170
|
0.006
|
NM_001161521
NM_001161522
|
KLHL2
|
kelch-like 2, Mayven (Drosophila)
|
|
chr10_-_69455926
|
0.006
|
NM_013266
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3
|
|
chr7_-_18067445
|
0.004
|
NM_175886
|
PRPS1L1
|
phosphoribosyl pyrophosphate synthetase 1-like 1
|
|
chr9_+_27109138
|
0.004
|
NM_000459
|
TEK
|
TEK tyrosine kinase, endothelial
|
|
chr4_-_82126200
|
0.000
|
NM_006259
|
PRKG2
|
protein kinase, cGMP-dependent, type II
|