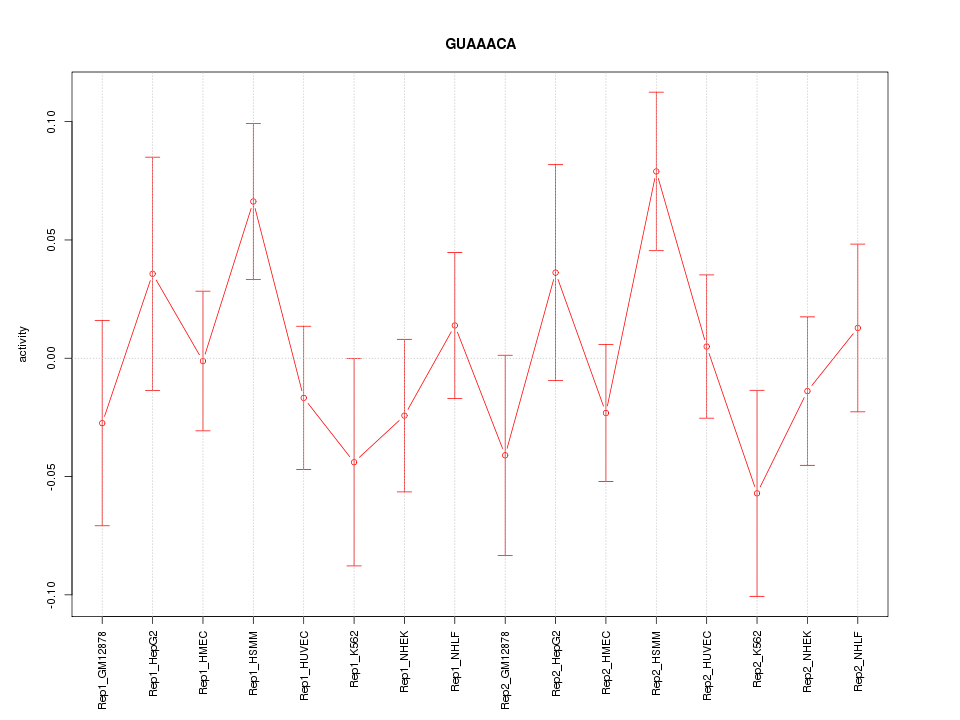
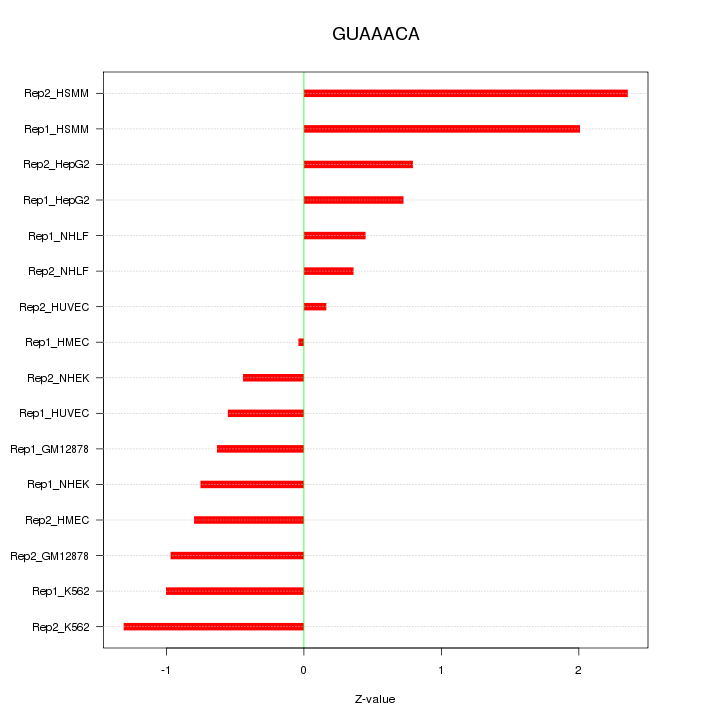
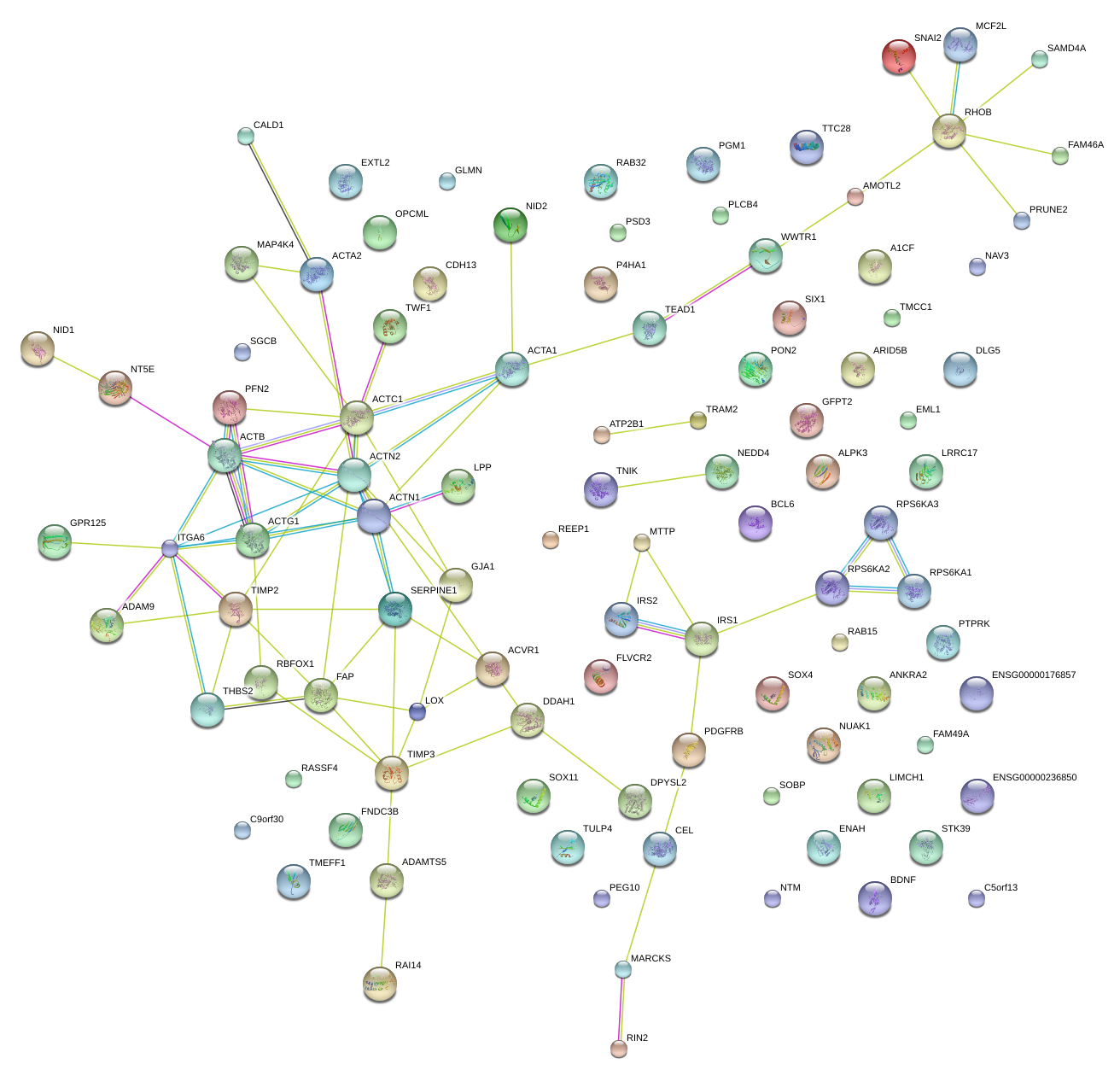
|
chr1_-_85930733
|
2.965
|
NM_012137
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr1_-_86043932
|
2.838
|
NM_001134445
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr7_+_102553343
|
2.594
|
NM_001031692
NM_005824
|
LRRC17
|
leucine rich repeat containing 17
|
|
chr15_-_35087748
|
2.438
|
NM_005159
|
ACTC1
|
actin, alpha, cardiac muscle 1
|
|
chr5_-_121412805
|
2.424
|
NM_001178102
|
LOX
|
lysyl oxidase
|
|
chr5_-_121414011
|
1.889
|
NM_002317
|
LOX
|
lysyl oxidase
|
|
chr2_+_20646831
|
1.875
|
NM_004040
|
RHOB
|
ras homolog gene family, member B
|
|
chr21_-_28339405
|
1.824
|
NM_007038
|
ADAMTS5
|
ADAM metallopeptidase with thrombospondin type 1 motif, 5
|
|
chr2_-_169103886
|
1.727
|
NM_013233
|
STK39
|
serine threonine kinase 39
|
|
chr3_-_149688638
|
1.699
|
NM_002628
NM_053024
|
PFN2
|
profilin 2
|
|
chr22_+_33196801
|
1.661
|
NM_000362
|
TIMP3
|
TIMP metallopeptidase inhibitor 3
|
|
chr7_+_100770378
|
1.653
|
NM_000602
NM_001165413
|
SERPINE1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1
|
|
chr6_+_86159301
|
1.565
|
NM_001204813
NM_002526
|
NT5E
|
5'-nucleotidase, ecto (CD73)
|
|
chr12_-_106533810
|
1.549
|
NM_014840
|
NUAK1
|
NUAK family, SNF1-like kinase, 1
|
|
chr2_-_227663454
|
1.549
|
NM_005544
|
IRS1
|
insulin receptor substrate 1
|
|
chr17_-_76921453
|
1.548
|
NM_003255
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr3_-_134093235
|
1.428
|
NM_016201
|
AMOTL2
|
angiomotin like 2
|
|
chr1_-_236228476
|
1.383
|
NM_002508
|
NID1
|
nidogen 1
|
|
chr6_-_82462374
|
1.317
|
NM_017633
|
FAM46A
|
family with sequence similarity 46, member A
|
|
chr1_-_225840660
|
1.315
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr8_-_18871163
|
1.304
|
NM_015310
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr5_-_111312522
|
1.200
|
NM_001142474
NM_001142475
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr12_+_78225068
|
1.180
|
NM_014903
|
NAV3
|
neuron navigator 3
|
|
chr10_+_63661012
|
1.170
|
NM_032199
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like)
|
|
chr8_-_18666295
|
1.170
|
NM_206909
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr6_-_128841432
|
1.127
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chr8_+_26435339
|
1.127
|
NM_001386
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr5_-_111093030
|
1.074
|
NM_001142482
NM_001142481
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr7_+_134464161
|
1.073
|
NM_004342
NM_033138
NM_033157
|
CALD1
|
caldesmon 1
|
|
chr5_-_111093792
|
1.067
|
NM_001142477
NM_001142476
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr15_+_85359910
|
1.024
|
NM_020778
|
ALPK3
|
alpha-kinase 3
|
|
chr5_-_111092918
|
0.985
|
NM_004772
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr8_+_26371461
|
0.967
|
NM_001197293
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr9_-_79520878
|
0.958
|
NM_015225
|
PRUNE2
|
prune homolog 2 (Drosophila)
|
|
chr8_+_38854431
|
0.957
|
NM_003816
|
ADAM9
|
ADAM metallopeptidase domain 9
|
|
chr20_+_9288446
|
0.957
|
NM_000933
|
PLCB4
|
phospholipase C, beta 4
|
|
chr2_-_16847070
|
0.956
|
NM_030797
|
FAM49A
|
family with sequence similarity 49, member A
|
|
chr14_-_69446038
|
0.954
|
NM_001102
NM_001130004
NM_001130005
|
ACTN1
|
actinin, alpha 1
|
|
chr20_+_9198036
|
0.946
|
NM_182797
|
PLCB4
|
phospholipase C, beta 4
|
|
chr4_+_100485239
|
0.944
|
NM_000253
|
MTTP
|
microsomal triglyceride transfer protein
|
|
chr11_+_12695851
|
0.941
|
NM_021961
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor)
|
|
chr20_+_9049660
|
0.934
|
NM_001172646
|
PLCB4
|
phospholipase C, beta 4
|
|
chr5_-_111093251
|
0.930
|
NM_001142479
NM_001142480
NM_001142478
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr5_-_179780314
|
0.924
|
NM_005110
|
GFPT2
|
glutamine-fructose-6-phosphate transaminase 2
|
|
chr4_-_52904424
|
0.917
|
NM_000232
|
SGCB
|
sarcoglycan, beta (43kDa dystrophin-associated glycoprotein)
|
|
chr3_-_187463474
|
0.910
|
NM_001706
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr14_-_61115906
|
0.907
|
NM_005982
|
SIX1
|
SIX homeobox 1
|
|
chr5_-_111091947
|
0.906
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr11_-_27742224
|
0.896
|
NM_001143805
NM_001143806
NM_170732
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr11_-_27721179
|
0.890
|
NM_170734
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr14_-_65438794
|
0.884
|
NM_198686
|
RAB15
|
RAB15, member RAS onocogene family
|
|
chr4_+_41614911
|
0.877
|
NM_001112719
NM_001112720
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr3_-_149421059
|
0.875
|
NM_001168278
|
WWTR1
|
WW domain containing transcription regulator 1
|
|
chr11_-_27722599
|
0.870
|
NM_001143808
NM_001143809
NM_001143810
NM_001143811
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr11_-_27722446
|
0.869
|
NM_001143812
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr2_+_173292306
|
0.865
|
NM_000210
NM_001079818
|
ITGA6
|
integrin, alpha 6
|
|
chr12_-_90049818
|
0.853
|
NM_001001323
NM_001682
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1
|
|
chr3_-_149375584
|
0.852
|
NM_001168280
NM_015472
|
WWTR1
|
WW domain containing transcription regulator 1
|
|
chr11_-_27681195
|
0.844
|
NM_001143816
NM_170735
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr2_-_86565205
|
0.839
|
NM_001164730
|
REEP1
|
receptor accessory protein 1
|
|
chr6_-_169654047
|
0.825
|
NM_003247
|
THBS2
|
thrombospondin 2
|
|
chr11_-_27741192
|
0.812
|
NM_001143807
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr7_+_134576150
|
0.808
|
NM_033139
NM_033140
|
CALD1
|
caldesmon 1
|
|
chr9_+_103204187
|
0.807
|
NM_001198812
|
C9orf30-TMEFF1
C9orf30
|
C9orf30-TMEFF1 readthrough
chromosome 9 open reading frame 30
|
|
chr2_-_163099877
|
0.806
|
NM_004460
|
FAP
|
fibroblast activation protein, alpha
|
|
chr5_+_34656595
|
0.805
|
NM_001145520
|
RAI14
|
retinoic acid induced 14
|
|
chr5_-_149535420
|
0.802
|
NM_002609
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr5_+_34684611
|
0.792
|
NM_001145521
NM_001145525
|
RAI14
|
retinoic acid induced 14
|
|
chr5_+_34687663
|
0.791
|
NM_001145523
|
RAI14
|
retinoic acid induced 14
|
|
chr16_+_82660390
|
0.787
|
NM_001220488
NM_001220489
NM_001220490
NM_001220491
NM_001220492
NM_001257
|
CDH13
|
cadherin 13, H-cadherin (heart)
|
|
chr13_+_113623496
|
0.783
|
NM_001112732
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr2_-_158732341
|
0.781
|
NM_001111067
|
ACVR1
|
activin A receptor, type I
|
|
chr4_+_41362750
|
0.779
|
NM_001112717
NM_001112718
NM_014988
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr11_-_27721975
|
0.772
|
NM_001143813
NM_001143814
NM_001709
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr11_-_27743604
|
0.766
|
NM_170731
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr11_-_27723061
|
0.763
|
NM_170733
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr7_-_95064213
|
0.762
|
NM_000305
NM_001018161
|
PON2
|
paraoxonase 2
|
|
chr5_+_34656367
|
0.758
|
NM_001145522
NM_015577
|
RAI14
|
retinoic acid induced 14
|
|
chr3_-_187454247
|
0.754
|
NM_001130845
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr11_+_131240370
|
0.752
|
NM_001048209
|
NTM
|
neurotrimin
|
|
chr13_-_110438896
|
0.745
|
NM_003749
|
IRS2
|
insulin receptor substrate 2
|
|
chr15_-_56285732
|
0.738
|
NM_006154
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr4_-_22517339
|
0.731
|
NM_145290
|
GPR125
|
G protein-coupled receptor 125
|
|
chr1_+_64058891
|
0.727
|
NM_002633
|
PGM1
|
phosphoglucomutase 1
|
|
chr3_-_187452669
|
0.691
|
NM_001134738
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr3_+_187943192
|
0.690
|
NM_001167671
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr14_+_76071804
|
0.685
|
NM_001195283
|
FLVCR2
|
feline leukemia virus subgroup C cellular receptor family, member 2
|
|
chr22_-_29075708
|
0.683
|
NM_001145418
|
TTC28
|
tetratricopeptide repeat domain 28
|
|
chr6_-_52441815
|
0.679
|
NM_012288
|
TRAM2
|
translocation associated membrane protein 2
|
|
chr1_+_64088886
|
0.674
|
NM_001172818
|
PGM1
|
phosphoglucomutase 1
|
|
chr3_+_187871473
|
0.669
|
NM_001167672
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr6_-_167275657
|
0.663
|
NM_001006932
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2
|
|
chr3_-_129599220
|
0.659
|
NM_001017395
|
TMCC1
|
transmembrane and coiled-coil domain family 1
|
|
chr3_+_187930720
|
0.652
|
NM_005578
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr10_+_45455149
|
0.642
|
NM_032023
|
RASSF4
|
Ras association (RalGDS/AF-6) domain family member 4
|
|
chr1_+_64059479
|
0.637
|
NM_001172819
|
PGM1
|
phosphoglucomutase 1
|
|
chr16_+_6823809
|
0.626
|
NM_001142334
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr3_+_171757413
|
0.622
|
NM_022763
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr2_+_102314162
|
0.617
|
NM_001242560
NM_004834
NM_145686
NM_001242559
NM_145687
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr13_+_113656027
|
0.609
|
NM_024979
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr16_+_6069094
|
0.607
|
NM_001142333
NM_018723
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr6_+_121756744
|
0.604
|
NM_000165
|
GJA1
|
gap junction protein, alpha 1, 43kDa
|
|
chr1_-_101360405
|
0.603
|
NM_001033025
NM_001439
|
EXTL2
|
exostoses (multiple)-like 2
|
|
chr6_+_158733691
|
0.596
|
NM_001007466
NM_020245
|
TULP4
|
tubby like protein 4
|
|
chr2_-_158731622
|
0.595
|
NM_001105
|
ACVR1
|
activin A receptor, type I
|
|
chr2_-_86564632
|
0.594
|
NM_001164731
NM_001164732
NM_022912
|
REEP1
|
receptor accessory protein 1
|
|
chr14_+_76044939
|
0.586
|
NM_017791
|
FLVCR2
|
feline leukemia virus subgroup C cellular receptor family, member 2
|
|
chr16_+_7382750
|
0.579
|
NM_145891
NM_145892
NM_145893
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr6_-_167040700
|
0.576
|
NM_021135
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2
|
|
chr8_-_49833823
|
0.574
|
NM_003068
|
SNAI2
|
snail homolog 2 (Drosophila)
|
|
chr9_+_103235489
|
0.570
|
NM_003692
|
TMEFF1
|
transmembrane protein with EGF-like and two follistatin-like domains 1
|
|
chr6_+_146864827
|
0.566
|
NM_006834
|
RAB32
|
RAB32, member RAS oncogene family
|
|
chr10_-_74856656
|
0.565
|
NM_000917
NM_001017962
NM_001142595
NM_001142596
|
P4HA1
|
prolyl 4-hydroxylase, alpha polypeptide I
|
|
chr7_+_94285620
|
0.564
|
NM_001040152
NM_001184961
NM_001184962
NM_015068
|
PEG10
|
paternally expressed 10
|
|
chr14_+_100259445
|
0.564
|
NM_001008707
NM_004434
|
EML1
|
echinoderm microtubule associated protein like 1
|
|
chr6_+_107811272
|
0.562
|
NM_018013
|
SOBP
|
sine oculis binding protein homolog (Drosophila)
|
|
chr14_-_52535945
|
0.548
|
NM_007361
|
NID2
|
nidogen 2 (osteonidogen)
|
|
chr10_-_79686279
|
0.546
|
NM_004747
|
DLG5
|
discs, large homolog 5 (Drosophila)
|
|
chr12_-_44200047
|
0.537
|
NM_001242397
NM_002822
|
TWF1
|
twinfilin, actin-binding protein, homolog 1 (Drosophila)
|
|
chr5_-_72861376
|
0.537
|
NM_023039
|
ANKRA2
|
ankyrin repeat, family A (RFXANK-like), 2
|
|
chr14_+_55034329
|
0.535
|
NM_001161576
NM_015589
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr2_+_5832778
|
0.532
|
NM_003108
|
SOX11
|
SRY (sex determining region Y)-box 11
|
|
chr6_+_114178516
|
0.532
|
NM_002356
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate
|
|
chr10_-_52645430
|
0.528
|
NM_001198818
NM_001198819
NM_001198820
NM_014576
NM_138932
NM_138933
|
A1CF
|
APOBEC1 complementation factor
|
|
chr3_-_46037299
|
0.525
|
NM_024513
|
FYCO1
|
FYVE and coiled-coil domain containing 1
|
|
chr11_+_112831968
|
0.525
|
NM_000615
NM_001076682
NM_001242607
NM_001242608
NM_181351
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr10_-_81205091
|
0.524
|
NM_153367
|
ZCCHC24
|
zinc finger, CCHC domain containing 24
|
|
chr7_+_94285676
|
0.523
|
NM_001172437
NM_001172438
|
PEG10
|
paternally expressed 10
|
|
chr20_+_61448391
|
0.513
|
NM_001853
|
COL9A3
|
collagen, type IX, alpha 3
|
|
chr17_+_53342320
|
0.502
|
NM_002126
|
HLF
|
hepatic leukemia factor
|
|
chr8_+_26240522
|
0.499
|
NM_004331
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like
|
|
chr6_-_52926588
|
0.495
|
NM_014920
NM_016513
|
ICK
|
intestinal cell (MAK-like) kinase
|
|
chrX_-_92928566
|
0.489
|
NM_004538
|
NAP1L3
|
nucleosome assembly protein 1-like 3
|
|
chr4_+_146403955
|
0.488
|
NM_001003688
|
SMAD1
|
SMAD family member 1
|
|
chr4_-_89744154
|
0.476
|
NM_001015045
|
FAM13A
|
family with sequence similarity 13, member A
|
|
chr14_-_39572414
|
0.473
|
NM_006364
|
SEC23A
|
Sec23 homolog A (S. cerevisiae)
|
|
chr15_-_78369825
|
0.471
|
NM_015079
NM_144572
|
TBC1D2B
|
TBC1 domain family, member 2B
|
|
chr8_+_58906968
|
0.464
|
NM_147189
|
FAM110B
|
family with sequence similarity 110, member B
|
|
chr4_-_89978117
|
0.462
|
NM_014883
|
FAM13A
|
family with sequence similarity 13, member A
|
|
chr7_+_94539209
|
0.460
|
NM_001166161
NM_001166162
NM_001166163
|
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A
|
|
chr17_-_41174431
|
0.457
|
NM_006373
|
VAT1
|
vesicle amine transport protein 1 homolog (T. californica)
|
|
chr4_-_16085593
|
0.457
|
NM_001145847
NM_001145848
|
PROM1
|
prominin 1
|
|
chr4_+_146402704
|
0.453
|
NM_005900
|
SMAD1
|
SMAD family member 1
|
|
chr3_-_57113292
|
0.449
|
NM_001128615
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr11_+_131780537
|
0.449
|
NM_001144058
NM_001144059
NM_016522
|
NTM
|
neurotrimin
|
|
chr7_+_128379345
|
0.449
|
NM_001130674
NM_001199671
NM_001199672
NM_001199673
NM_001199674
NM_001219
|
CALU
|
calumenin
|
|
chr3_-_129612371
|
0.448
|
NM_001128224
|
TMCC1
|
transmembrane and coiled-coil domain family 1
|
|
chrX_-_131623994
|
0.446
|
NM_001170704
|
MBNL3
|
muscleblind-like 3 (Drosophila)
|
|
chr2_+_62900985
|
0.444
|
NM_001142615
|
EHBP1
|
EH domain binding protein 1
|
|
chr7_-_35077652
|
0.442
|
NM_015283
|
DPY19L1
|
dpy-19-like 1 (C. elegans)
|
|
chr10_+_116853118
|
0.439
|
NM_207303
|
ATRNL1
|
attractin-like 1
|
|
chr16_-_66959418
|
0.438
|
NM_001128850
NM_004165
|
RRAD
|
Ras-related associated with diabetes
|
|
chr15_-_56209305
|
0.434
|
NM_198400
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr5_-_72744351
|
0.433
|
NM_004472
|
FOXD1
|
forkhead box D1
|
|
chr4_-_16077740
|
0.430
|
NM_001145849
NM_001145850
NM_001145851
NM_001145852
NM_006017
|
PROM1
|
prominin 1
|
|
chr1_-_84464728
|
0.428
|
NM_024686
|
TTLL7
|
tubulin tyrosine ligase-like family, member 7
|
|
chr10_+_112836784
|
0.423
|
NM_000681
|
ADRA2A
|
adrenergic, alpha-2A-, receptor
|
|
chrX_+_37208527
|
0.423
|
NM_000950
NM_001142395
NM_001173486
NM_001173489
NM_001173490
|
PRRG1
|
proline rich Gla (G-carboxyglutamic acid) 1
|
|
chr9_-_107690526
|
0.422
|
NM_005502
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr3_-_52719514
|
0.420
|
NM_018313
|
PBRM1
|
polybromo 1
|
|
chr17_+_42634598
|
0.417
|
NM_001466
|
FZD2
|
frizzled family receptor 2
|
|
chrX_-_131547536
|
0.412
|
NM_001170701
NM_001170702
NM_001170703
|
MBNL3
|
muscleblind-like 3 (Drosophila)
|
|
chr16_+_69599868
|
0.404
|
NM_001113178
NM_006599
NM_138713
NM_138714
NM_173214
NM_173215
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr8_+_67687415
|
0.397
|
NM_013257
NM_170709
|
SGK3
|
serum/glucocorticoid regulated kinase family, member 3
|
|
chr2_+_118846029
|
0.394
|
NM_016133
|
INSIG2
|
insulin induced gene 2
|
|
chr18_+_55862577
|
0.393
|
NM_001144970
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr19_+_57702833
|
0.393
|
NM_003417
|
ZNF264
|
zinc finger protein 264
|
|
chr15_+_69452799
|
0.388
|
NM_015554
|
GLCE
|
glucuronic acid epimerase
|
|
chr17_-_2304217
|
0.388
|
NM_020310
|
MNT
|
MAX binding protein
|
|
chr9_-_23821842
|
0.374
|
NM_001171195
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr9_-_16870719
|
0.373
|
NM_017637
|
BNC2
|
basonuclin 2
|
|
chr4_-_186877869
|
0.373
|
NM_021069
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr1_+_192778168
|
0.372
|
NM_002923
|
RGS2
|
regulator of G-protein signaling 2, 24kDa
|
|
chr11_+_125034558
|
0.372
|
NM_022062
|
PKNOX2
|
PBX/knotted 1 homeobox 2
|
|
chr17_+_70117154
|
0.370
|
NM_000346
|
SOX9
|
SRY (sex determining region Y)-box 9
|
|
chr7_+_94536926
|
0.369
|
NM_001166160
NM_017650
|
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A
|
|
chr4_-_186732208
|
0.369
|
NM_001145671
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr10_-_101945787
|
0.364
|
NM_001100626
|
ERLIN1
|
ER lipid raft associated 1
|
|
chr2_+_62932786
|
0.356
|
NM_001142616
NM_015252
|
EHBP1
|
EH domain binding protein 1
|
|
chr7_-_27224762
|
0.354
|
NM_005523
|
HOXA11
|
homeobox A11
|
|
chr4_-_186732047
|
0.353
|
NM_001145672
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr4_-_186733372
|
0.350
|
NM_003603
NM_001145670
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr8_-_131455833
|
0.349
|
NM_001247996
NM_018482
|
ASAP1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1
|
|
chr14_+_59104736
|
0.347
|
NM_001079520
NM_016651
|
DACT1
|
dapper, antagonist of beta-catenin, homolog 1 (Xenopus laevis)
|
|
chr3_-_56809594
|
0.347
|
NM_001128616
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr3_+_171758289
|
0.344
|
NM_001135095
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr12_-_31743822
|
0.343
|
NM_144973
|
DENND5B
|
DENN/MADD domain containing 5B
|
|
chr6_-_83903632
|
0.342
|
NM_001199917
|
PGM3
|
phosphoglucomutase 3
|
|
chr18_-_48724031
|
0.341
|
NM_016626
|
MEX3C
|
mex-3 homolog C (C. elegans)
|
|
chr4_-_186697065
|
0.340
|
NM_001145673
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr2_-_167232464
|
0.338
|
NM_002977
|
SCN9A
|
sodium channel, voltage-gated, type IX, alpha subunit
|
|
chr5_-_58334936
|
0.337
|
NM_001197221
NM_001197222
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr10_-_101945686
|
0.337
|
NM_006459
|
ERLIN1
|
ER lipid raft associated 1
|
|
chr6_+_108882068
|
0.336
|
NM_001455
|
FOXO3
|
forkhead box O3
|
|
chr9_-_73029539
|
0.333
|
NM_001206
|
KLF9
|
Kruppel-like factor 9
|
|
chr8_-_103667882
|
0.332
|
NM_005655
|
KLF10
|
Kruppel-like factor 10
|
|
chr15_+_99192696
|
0.332
|
NM_000875
|
IGF1R
|
insulin-like growth factor 1 receptor
|
|
chr8_+_9413439
|
0.332
|
NM_003747
|
TNKS
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase
|
|
chr9_+_4679557
|
0.323
|
NM_017913
|
CDC37L1
|
cell division cycle 37 homolog (S. cerevisiae)-like 1
|
|
chr12_-_80084742
|
0.323
|
NM_002583
|
PAWR
|
PRKC, apoptosis, WT1, regulator
|