|
chrX_+_43514157
|
2.646
|
|
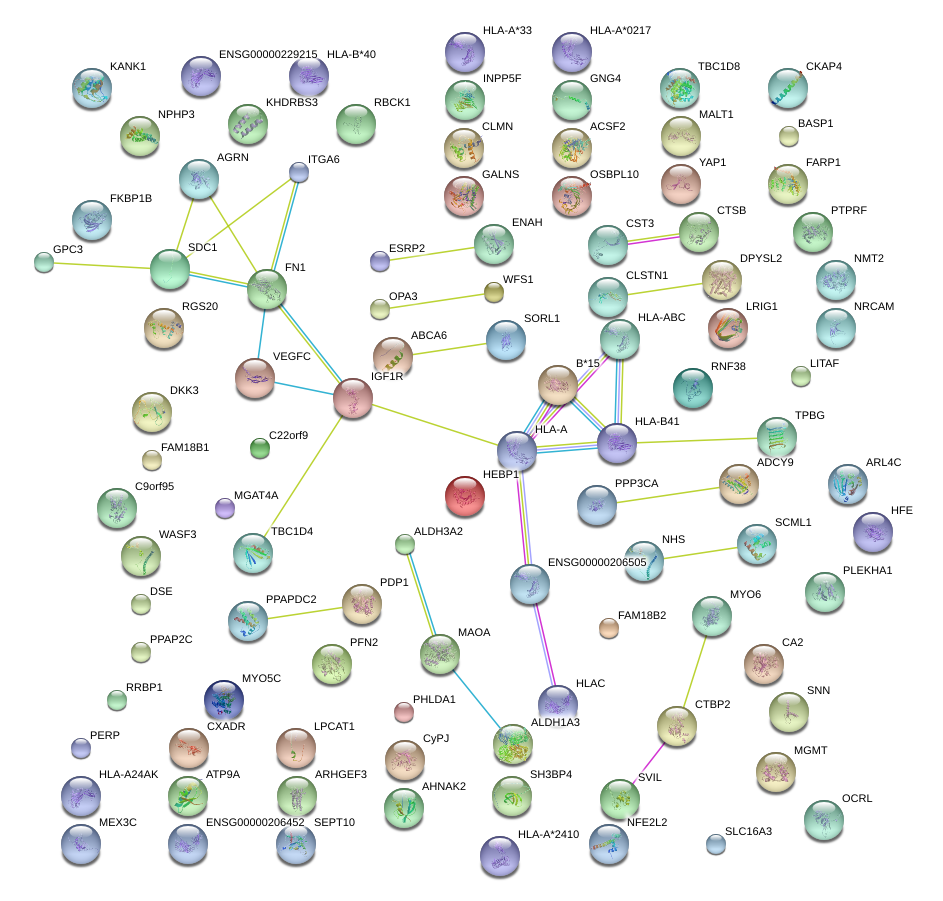
MAOA
|
monoamine oxidase A
|
|
chr2_-_99347430
|
1.966
|
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A
|
|
chr8_+_136469557
|
1.665
|
NM_006558
|
KHDRBS3
|
KH domain containing, RNA binding, signal transduction associated 3
|
|
chr5_+_17217781
|
1.640
|
|
BASP1
|
brain abundant, membrane attached signal protein 1
|
|
chr5_+_17217474
|
1.449
|
NM_006317
|
BASP1
|
brain abundant, membrane attached signal protein 1
|
|
chr2_+_235860616
|
1.387
|
NM_014521
|
SH3BP4
|
SH3-domain binding protein 4
|
|
chr13_+_27131879
|
1.346
|
|
WASF3
|
WAS protein family, member 3
|
|
chr21_+_18885296
|
1.294
|
|
CXADR
|
coxsackie virus and adenovirus receptor
|
|
chr8_+_26371461
|
1.244
|
NM_001197293
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr22_-_45636760
|
1.175
|
|
KIAA0930
|
KIAA0930
|
|
chr15_+_101420004
|
1.163
|
NM_000693
|
ALDH1A3
|
aldehyde dehydrogenase 1 family, member A3
|
|
chr3_-_149688886
|
1.116
|
|
PFN2
|
profilin 2
|
|
chr2_-_110371669
|
1.115
|
NM_144710
NM_178584
|
SEPT10
|
septin 10
|
|
chr2_-_20424627
|
1.064
|
NM_002997
|
SDC1
|
syndecan 1
|
|
chr3_-_149688638
|
1.058
|
NM_002628
NM_053024
|
PFN2
|
profilin 2
|
|
chr11_+_121322848
|
1.031
|
NM_003105
|
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing
|
|
chr2_-_99347588
|
1.024
|
NM_012214
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A
|
|
chr20_-_23618382
|
1.008
|
NM_000099
|
CST3
|
cystatin C
|
|
chr20_+_388941
|
0.989
|
|
RBCK1
|
RanBP-type and C3HC4-type zinc finger containing 1
|
|
chr10_+_131265500
|
0.944
|
|
MGMT
|
O-6-methylguanine-DNA methyltransferase
|
|
chr2_-_20425166
|
0.943
|
NM_001006946
|
SDC1
|
syndecan 1
|
|
chr14_-_95786095
|
0.937
|
NM_024734
|
CLMN
|
calmin (calponin-like, transmembrane)
|
|
chr21_+_18885104
|
0.929
|
NM_001207063
NM_001207064
NM_001207065
NM_001207066
NM_001338
|
CXADR
|
coxsackie virus and adenovirus receptor
|
|
chr1_-_9884024
|
0.921
|
|
CLSTN1
|
calsyntenin 1
|
|
chr6_+_83073954
|
0.909
|
NM_001166392
|
TPBG
|
trophoblast glycoprotein
|
|
chr15_-_52587851
|
0.896
|
NM_018728
|
MYO5C
|
myosin VC
|
|
chrX_-_133119624
|
0.881
|
NM_001164617
NM_001164618
NM_001164619
NM_004484
|
GPC3
|
glypican 3
|
|
chr6_+_29855349
|
0.857
|
|
HLA-H
|
major histocompatibility complex, class I, H (pseudogene)
|
|
chr15_+_101420032
|
0.856
|
|
ALDH1A3
|
aldehyde dehydrogenase 1 family, member A3
|
|
chr14_-_105444693
|
0.831
|
NM_138420
|
AHNAK2
|
AHNAK nucleoprotein 2
|
|
chr2_-_101767714
|
0.819
|
NM_001102426
|
TBC1D8
|
TBC1 domain family, member 8 (with GRAM domain)
|
|
chr1_-_9884549
|
0.807
|
NM_001009566
NM_014944
|
CLSTN1
|
calsyntenin 1
|
|
chr6_-_138428477
|
0.800
|
NM_022121
|
PERP
|
PERP, TP53 apoptosis effector
|
|
chr10_+_124134093
|
0.792
|
NM_001001974
|
PLEKHA1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1
|
|
chrX_-_133119475
|
0.783
|
|
GPC3
|
glypican 3
|
|
chr12_-_76425207
|
0.770
|
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chr10_-_30024594
|
0.765
|
NM_003174
|
SVIL
|
supervillin
|
|
chr18_-_48724031
|
0.759
|
NM_016626
|
MEX3C
|
mex-3 homolog C (C. elegans)
|
|
chr17_+_80186281
|
0.759
|
NM_001042422
NM_001206950
|
SLC16A3
|
solute carrier family 16, member 3 (monocarboxylic acid transporter 4)
|
|
chr15_+_99191671
|
0.737
|
|
IGF1R
|
insulin-like growth factor 1 receptor
|
|
chr5_-_1524022
|
0.732
|
NM_024830
|
LPCAT1
|
lysophosphatidylcholine acyltransferase 1
|
|
chr6_+_29910308
|
0.731
|
|
HLA-A
|
major histocompatibility complex, class I, A
|
|
chr2_-_235405692
|
0.719
|
NM_005737
|
ARL4C
|
ADP-ribosylation factor-like 4C
|
|
chr11_-_12030539
|
0.711
|
|
DKK3
|
dickkopf 3 homolog (Xenopus laevis)
|
|
chr17_+_48503607
|
0.710
|
|
ACSF2
|
acyl-CoA synthetase family member 2
|
|
chr20_+_388963
|
0.709
|
|
RBCK1
|
RanBP-type and C3HC4-type zinc finger containing 1
|
|
chr10_-_126849556
|
0.697
|
NM_001083914
|
CTBP2
|
C-terminal binding protein 2
|
|
chr12_-_106641675
|
0.694
|
NM_006825
|
CKAP4
|
cytoskeleton-associated protein 4
|
|
chr3_-_57113292
|
0.691
|
NM_001128615
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr20_-_50384861
|
0.689
|
|
ATP9A
|
ATPase, class II, type 9A
|
|
chr4_-_102268626
|
0.680
|
NM_000944
NM_001130691
NM_001130692
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme
|
|
chr1_-_235812994
|
0.679
|
NM_001098722
|
GNG4
|
guanine nucleotide binding protein (G protein), gamma 4
|
|
chr6_+_29855506
|
0.673
|
|
|
|
|
chr11_-_12030622
|
0.659
|
NM_015881
|
DKK3
|
dickkopf 3 homolog (Xenopus laevis)
|
|
chr7_-_108096693
|
0.659
|
NM_001193582
NM_001193583
NM_001193584
NM_005010
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr1_+_43996478
|
0.652
|
NM_002840
NM_130440
|
PTPRF
|
protein tyrosine phosphatase, receptor type, F
|
|
chr11_+_101980736
|
0.649
|
NM_001130145
NM_001195044
NM_006106
|
YAP1
|
Yes-associated protein 1
|
|
chr8_+_94929082
|
0.640
|
NM_018444
NM_001161780
NM_001161779
NM_001161781
|
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1
|
|
chr16_-_11680756
|
0.632
|
NM_001136472
NM_001136473
|
LITAF
|
lipopolysaccharide-induced TNF factor
|
|
chr10_+_131265453
|
0.625
|
NM_002412
|
MGMT
|
O-6-methylguanine-DNA methyltransferase
|
|
chr19_-_291168
|
0.625
|
NM_177543
|
PPAP2C
|
phosphatidic acid phosphatase type 2C
|
|
chr16_+_11762235
|
0.619
|
NM_003498
|
SNN
|
stannin
|
|
chr20_+_388717
|
0.618
|
|
RBCK1
|
RanBP-type and C3HC4-type zinc finger containing 1
|
|
chr9_+_504661
|
0.618
|
NM_015158
|
KANK1
|
KN motif and ankyrin repeat domains 1
|
|
chr10_-_15210610
|
0.617
|
NM_004808
|
NMT2
|
N-myristoyltransferase 2
|
|
chr10_-_126849010
|
0.598
|
|
CTBP2
|
C-terminal binding protein 2
|
|
chr22_-_45636584
|
0.591
|
NM_001009880
|
KIAA0930
|
KIAA0930
|
|
chr13_+_27131799
|
0.589
|
NM_006646
|
WASF3
|
WAS protein family, member 3
|
|
chr18_+_56338783
|
0.589
|
|
MALT1
|
mucosa associated lymphoid tissue lymphoma translocation gene 1
|
|
chr13_+_98795468
|
0.588
|
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived)
|
|
chr9_-_36400817
|
0.587
|
NM_194328
NM_194332
|
RNF38
|
ring finger protein 38
|
|
chr2_-_216300770
|
0.584
|
NM_002026
NM_054034
NM_212474
NM_212476
NM_212478
NM_212482
|
FN1
|
fibronectin 1
|
|
chr16_-_68269948
|
0.580
|
NM_024939
|
ESRP2
|
epithelial splicing regulatory protein 2
|
|
chr1_+_955468
|
0.580
|
NM_198576
|
AGRN
|
agrin
|
|
chr10_-_126849066
|
0.579
|
NM_001329
|
CTBP2
|
C-terminal binding protein 2
|
|
chr8_+_86376130
|
0.579
|
NM_000067
|
CA2
|
carbonic anhydrase II
|
|
chrX_+_17755562
|
0.569
|
NM_001037535
NM_001037536
NM_001037540
NM_006746
|
SCML1
|
sex comb on midleg-like 1 (Drosophila)
|
|
chr9_-_36401149
|
0.566
|
NM_194330
|
RNF38
|
ring finger protein 38
|
|
chr17_-_15466772
|
0.565
|
NM_001204478
NM_001135036
NM_145301
|
FAM18B2-CDRT4
FAM18B2
|
FAM18B2-CDRT4 readthrough
family with sequence similarity 18, member B2
|
|
chr4_+_6271605
|
0.563
|
|
WFS1
|
Wolfram syndrome 1 (wolframin)
|
|
chr10_+_121485638
|
0.562
|
|
INPP5F
|
inositol polyphosphate-5-phosphatase F
|
|
chr6_+_116692098
|
0.559
|
NM_013352
|
DSE
|
dermatan sulfate epimerase
|
|
chr12_-_76425375
|
0.558
|
NM_007350
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chr13_-_76055872
|
0.557
|
NM_014832
|
TBC1D4
|
TBC1 domain family, member 4
|
|
chr2_+_173292306
|
0.551
|
NM_000210
NM_001079818
|
ITGA6
|
integrin, alpha 6
|
|
chr10_+_124134206
|
0.549
|
|
PLEKHA1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1
|
|
chr8_-_11725509
|
0.547
|
|
CTSB
|
cathepsin B
|
|
chr16_-_4166185
|
0.543
|
NM_001116
|
ADCY9
|
adenylate cyclase 9
|
|
chr1_-_225840660
|
0.538
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr3_-_32023237
|
0.536
|
NM_001174060
NM_017784
|
OSBPL10
|
oxysterol binding protein-like 10
|
|
chr3_-_132441216
|
0.532
|
|
NPHP3
|
nephronophthisis 3 (adolescent)
|
|
chr20_-_50384899
|
0.528
|
NM_006045
|
ATP9A
|
ATPase, class II, type 9A
|
|
chr6_+_76458888
|
0.527
|
NM_004999
|
MYO6
|
myosin VI
|
|
chr16_-_88923233
|
0.520
|
|
GALNS
|
galactosamine (N-acetyl)-6-sulfate sulfatase
|
|
chr20_-_17662704
|
0.517
|
|
RRBP1
|
ribosome binding protein 1 homolog 180kDa (dog)
|
|
chr17_+_48503518
|
0.512
|
NM_025149
|
ACSF2
|
acyl-CoA synthetase family member 2
|
|
chr4_-_177713664
|
0.508
|
NM_005429
|
VEGFC
|
vascular endothelial growth factor C
|
|
chr20_-_17662828
|
0.508
|
NM_001042576
NM_004587
|
RRBP1
|
ribosome binding protein 1 homolog 180kDa (dog)
|
|
chr12_-_13153200
|
0.508
|
NM_015987
|
HEBP1
|
heme binding protein 1
|
|
chr6_+_76458970
|
0.505
|
|
MYO6
|
myosin VI
|
|
chr2_+_24272627
|
0.503
|
|
FKBP1B
|
FK506 binding protein 1B, 12.6 kDa
|
|
chr9_-_77703040
|
0.503
|
NM_001127603
NM_017881
|
C9orf95
|
chromosome 9 open reading frame 95
|
|
chr3_-_66551341
|
0.494
|
|
LRIG1
|
leucine-rich repeats and immunoglobulin-like domains 1
|
|
chr17_+_19551835
|
0.491
|
NM_000382
NM_001031806
|
ALDH3A2
|
aldehyde dehydrogenase 3 family, member A2
|
|
chr6_+_138188352
|
0.491
|
NM_006290
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr14_-_53417636
|
0.491
|
|
FERMT2
|
fermitin family member 2
|
|
chr19_-_291335
|
0.490
|
NM_003712
NM_177526
|
PPAP2C
|
phosphatidic acid phosphatase type 2C
|
|
chr1_-_11714738
|
0.489
|
NM_012168
|
FBXO2
|
F-box protein 2
|
|
chr5_+_72921980
|
0.484
|
NM_001080479
NM_001177693
|
RGNEF
|
190 kDa guanine nucleotide exchange factor
|
|
chr7_-_38670988
|
0.483
|
NM_001635
NM_139316
|
AMPH
|
amphiphysin
|
|
chr7_-_38670950
|
0.480
|
|
AMPH
|
amphiphysin
|
|
chr9_+_33750463
|
0.466
|
NM_001197097
NM_007343
NM_001197098
|
PRSS3
|
protease, serine, 3
|
|
chr1_-_11714471
|
0.465
|
|
FBXO2
|
F-box protein 2
|
|
chr2_+_173292344
|
0.464
|
|
ITGA6
|
integrin, alpha 6
|
|
chr18_+_56338818
|
0.463
|
|
MALT1
|
mucosa associated lymphoid tissue lymphoma translocation gene 1
|
|
chr2_-_20212408
|
0.462
|
|
MATN3
|
matrilin 3
|
|
chr16_-_4588379
|
0.461
|
NM_001199054
|
C16orf5
|
chromosome 16 open reading frame 5
|
|
chr1_+_9352955
|
0.460
|
|
SPSB1
|
splA/ryanodine receptor domain and SOCS box containing 1
|
|
chr8_+_26240522
|
0.459
|
NM_004331
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like
|
|
chr4_-_7941092
|
0.456
|
NM_001134647
NM_198595
|
AFAP1
|
actin filament associated protein 1
|
|
chr2_+_238600780
|
0.453
|
NM_001137551
NM_001137552
NM_001137553
NM_004735
|
LRRFIP1
|
leucine rich repeat (in FLII) interacting protein 1
|
|
chr1_-_11714396
|
0.450
|
|
FBXO2
|
F-box protein 2
|
|
chr19_+_16187315
|
0.450
|
|
TPM4
|
tropomyosin 4
|
|
chr6_-_3752245
|
0.450
|
NM_183373
|
PXDC1
|
PX domain containing 1
|
|
chr2_-_20850822
|
0.447
|
NM_022460
|
HS1BP3
|
HCLS1 binding protein 3
|
|
chr17_+_80186886
|
0.447
|
NM_004207
|
SLC16A3
|
solute carrier family 16, member 3 (monocarboxylic acid transporter 4)
|
|
chr7_+_45927958
|
0.446
|
NM_000596
|
IGFBP1
|
insulin-like growth factor binding protein 1
|
|
chr11_-_5526833
|
0.444
|
NM_001005567
|
OR51B5
HBE1
|
olfactory receptor, family 51, subfamily B, member 5
hemoglobin, epsilon 1
|
|
chr2_+_238600878
|
0.444
|
|
LRRFIP1
|
leucine rich repeat (in FLII) interacting protein 1
|
|
chr15_+_39873276
|
0.444
|
NM_003246
|
THBS1
|
thrombospondin 1
|
|
chr19_+_38755422
|
0.444
|
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr10_+_121485593
|
0.442
|
|
INPP5F
|
inositol polyphosphate-5-phosphatase F
|
|
chr5_+_67511578
|
0.441
|
NM_181523
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr10_+_120863626
|
0.440
|
|
FAM45A
FAM45B
|
family with sequence similarity 45, member A
family with sequence similarity 45, member A pseudogene
|
|
chr9_-_6645564
|
0.436
|
NM_000170
|
GLDC
|
glycine dehydrogenase (decarboxylating)
|
|
chr21_+_42540054
|
0.435
|
|
BACE2
|
beta-site APP-cleaving enzyme 2
|
|
chr17_-_74497406
|
0.435
|
NM_024599
|
RHBDF2
|
rhomboid 5 homolog 2 (Drosophila)
|
|
chr10_+_121485487
|
0.434
|
NM_001243195
NM_014937
|
INPP5F
|
inositol polyphosphate-5-phosphatase F
|
|
chr10_-_91011516
|
0.426
|
|
LIPA
|
lipase A, lysosomal acid, cholesterol esterase
|
|
chr8_+_26435339
|
0.425
|
NM_001386
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr16_+_27325244
|
0.424
|
NM_000418
NM_001008699
|
IL4R
|
interleukin 4 receptor
|
|
chr19_+_13875345
|
0.423
|
|
MRI1
|
methylthioribose-1-phosphate isomerase homolog (S. cerevisiae)
|
|
chr20_+_6748744
|
0.413
|
NM_001200
|
BMP2
|
bone morphogenetic protein 2
|
|
chr2_-_110371404
|
0.412
|
|
SEPT10
|
septin 10
|
|
chr18_+_60190657
|
0.412
|
NM_017742
|
ZCCHC2
|
zinc finger, CCHC domain containing 2
|
|
chr14_-_53162418
|
0.409
|
NM_014584
|
ERO1L
|
ERO1-like (S. cerevisiae)
|
|
chr12_-_13153047
|
0.408
|
|
HEBP1
|
heme binding protein 1
|
|
chr12_-_80084023
|
0.406
|
|
PAWR
|
PRKC, apoptosis, WT1, regulator
|
|
chr22_+_31608249
|
0.406
|
NM_005569
|
LIMK2
|
LIM domain kinase 2
|
|
chr6_+_142622797
|
0.405
|
NM_001032394
NM_001032395
NM_020455
NM_198569
|
GPR126
|
G protein-coupled receptor 126
|
|
chr18_+_55711890
|
0.405
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr14_+_20937537
|
0.404
|
NM_000270
|
PNP
|
purine nucleoside phosphorylase
|
|
chr1_+_9352931
|
0.401
|
NM_025106
|
SPSB1
|
splA/ryanodine receptor domain and SOCS box containing 1
|
|
chr8_-_11725622
|
0.400
|
|
CTSB
|
cathepsin B
|
|
chr19_-_11373117
|
0.397
|
NM_020812
|
DOCK6
|
dedicator of cytokinesis 6
|
|
chr2_-_165477986
|
0.396
|
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chr5_+_156693051
|
0.394
|
NM_001037333
NM_001037332
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr1_-_95392501
|
0.393
|
NM_001839
|
CNN3
|
calponin 3, acidic
|
|
chr21_+_42540092
|
0.393
|
|
BACE2
|
beta-site APP-cleaving enzyme 2
|
|
chr1_+_35247838
|
0.393
|
NM_001005752
|
GJB3
|
gap junction protein, beta 3, 31kDa
|
|
chr3_+_171758289
|
0.390
|
NM_001135095
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chrX_+_49188092
|
0.388
|
NM_012196
NM_001127200
NM_001098412
|
GAGE8
GAGE2C
GAGE2E
GAGE13
GAGE2A
|
G antigen 8
G antigen 2C
G antigen 2E
G antigen 13
G antigen 2A
|
|
chr5_-_139726173
|
0.387
|
NM_001945
|
HBEGF
|
heparin-binding EGF-like growth factor
|
|
chr1_+_64058891
|
0.387
|
NM_002633
|
PGM1
|
phosphoglucomutase 1
|
|
chr8_-_11725587
|
0.386
|
|
CTSB
|
cathepsin B
|
|
chr5_-_179334850
|
0.384
|
NM_015043
NM_198868
|
TBC1D9B
|
TBC1 domain family, member 9B (with GRAM domain)
|
|
chrX_+_49216616
|
0.383
|
NM_012196
NM_001472
NM_001474
NM_001475
NM_021123
NM_001477
NM_001127200
NM_001127212
|
GAGE8
GAGE1
GAGE2C
GAGE4
GAGE5
GAGE6
GAGE7
GAGE12I
GAGE2E
GAGE12B
GAGE2A
|
G antigen 8
G antigen 1
G antigen 2C
G antigen 4
G antigen 5
G antigen 6
G antigen 7
G antigen 12I
G antigen 2E
G antigen 12B
G antigen 2A
|
|
chr2_+_177053306
|
0.382
|
NM_024501
|
HOXD1
|
homeobox D1
|
|
chrX_+_49207115
|
0.382
|
NM_012196
NM_001472
NM_001127200
NM_001098407
NM_001127212
|
GAGE8
GAGE2C
GAGE2E
GAGE2D
GAGE2A
|
G antigen 8
G antigen 2C
G antigen 2E
G antigen 2D
G antigen 2A
|
|
chr22_-_36783977
|
0.381
|
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr20_+_25228698
|
0.381
|
NM_002862
|
PYGB
|
phosphorylase, glycogen; brain
|
|
chr13_-_99229226
|
0.380
|
NM_001032296
|
STK24
|
serine/threonine kinase 24
|
|
chr17_+_42422665
|
0.380
|
|
GRN
|
granulin
|
|
chr11_+_706103
|
0.379
|
NM_022772
|
EPS8L2
|
EPS8-like 2
|
|
chr3_+_43328001
|
0.378
|
NM_001100594
NM_017719
|
SNRK
|
SNF related kinase
|
|
chr18_-_11148760
|
0.378
|
NM_022068
|
PIEZO2
|
piezo-type mechanosensitive ion channel component 2
|
|
chr22_-_50946129
|
0.376
|
NM_033200
|
LMF2
|
lipase maturation factor 2
|
|
chrX_+_49197555
|
0.375
|
NM_012196
NM_001472
NM_001127200
NM_001098407
NM_001127212
|
GAGE8
GAGE2C
GAGE2E
GAGE12J
GAGE2D
GAGE2A
|
G antigen 8
G antigen 2C
G antigen 2E
G antigen 12J
G antigen 2D
G antigen 2A
|
|
chr22_-_50946020
|
0.375
|
|
LMF2
|
lipase maturation factor 2
|
|
chr17_+_75137101
|
0.374
|
|
SEC14L1
|
SEC14-like 1 (S. cerevisiae)
|
|
chr16_-_88923348
|
0.373
|
NM_000512
|
GALNS
|
galactosamine (N-acetyl)-6-sulfate sulfatase
|
|
chr22_-_43045288
|
0.371
|
|
CYB5R3
|
cytochrome b5 reductase 3
|
|
chr14_-_74960035
|
0.370
|
NM_006432
|
NPC2
|
Niemann-Pick disease, type C2
|
|
chr6_-_134496833
|
0.369
|
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr10_-_101769640
|
0.368
|
NM_015221
|
DNMBP
|
dynamin binding protein
|
|
chr19_+_3094528
|
0.368
|
|
GNA11
|
guanine nucleotide binding protein (G protein), alpha 11 (Gq class)
|
|
chr15_-_52821004
|
0.367
|
NM_000259
NM_001142495
|
MYO5A
|
myosin VA (heavy chain 12, myoxin)
|
|
chr16_-_53537112
|
0.366
|
NM_001012398
NM_022476
|
AKTIP
|
AKT interacting protein
|
|
chr2_+_24272583
|
0.364
|
NM_004116
NM_054033
|
FKBP1B
|
FK506 binding protein 1B, 12.6 kDa
|
|
chr15_-_90358057
|
0.364
|
NM_001150
|
ANPEP
|
alanyl (membrane) aminopeptidase
|
|
chr1_+_87170515
|
0.364
|
|
SH3GLB1
|
SH3-domain GRB2-like endophilin B1
|
|
chr2_+_238601052
|
0.364
|
|
LRRFIP1
|
leucine rich repeat (in FLII) interacting protein 1
|
|
chr15_+_75494235
|
0.363
|
|
C15orf39
|
chromosome 15 open reading frame 39
|
|
chr22_-_36784001
|
0.362
|
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr19_-_7293876
|
0.361
|
NM_000208
NM_001079817
|
INSR
|
insulin receptor
|
|
chr3_+_105085553
|
0.360
|
NM_001243280
NM_001243281
NM_001243283
NM_001627
|
ALCAM
|
activated leukocyte cell adhesion molecule
|
|
chr22_-_43045379
|
0.360
|
NM_000398
|
CYB5R3
|
cytochrome b5 reductase 3
|
|
chr16_-_57318574
|
0.359
|
NM_015993
|
PLLP
|
plasmolipin
|
|
chr20_+_388693
|
0.358
|
NM_006462
NM_031229
|
RBCK1
|
RanBP-type and C3HC4-type zinc finger containing 1
|
|
chr5_-_127873734
|
0.358
|
NM_001999
|
FBN2
|
fibrillin 2
|